Original research
Concordance analysis between noninvasive prenatal testing
(NIPT) and prenatal karyotyping for detecting fetal aneuploidies
Análisis de concordancia entre la prueba prenatal no invasiva (NIPT) y el cariotipo prenatal para la detección de aneuploidías fetales
Diana Carolina Grajales-Ospina1 Luz Karime Yunis-Hazbun1,2
Luz Karime Yunis-Hazbun1,2 Andrés Mauricio Camacho-Montaño3
Andrés Mauricio Camacho-Montaño3 Alejandro Antonio Bautista-Charry4
Alejandro Antonio Bautista-Charry4 Reinaldo Niño-Alba4
Reinaldo Niño-Alba4 Juan José Yunis1,2,5,6
Juan José Yunis1,2,5,6
1 Universidad Nacional de Colombia - Bogotá Campus - Faculty of Medicine - Molecular Pathology Research Group - Bogotá D.C. - Colombia.
2 Servicios Médicos Yunis Turbay y Cia. SAS - Genetics Institute - Molecular Diagnostic Service - Bogotá D.C. - Colombia.
3 Subred Integrada de Servicios de Salud Centro Oriente Empresa Social del Estado - Gynecology and Obstetrics Service - Bogotá D.C. - Colombia.
4 Universidad Nacional de Colombia - Bogotá Campus - Faculty of Medicine - Obstetrics and Gynecology Department - Bogotá D.C. - Colombia.
5 Universidad Nacional de Colombia - Bogotá Campus - Faculty of Medicine - Department of Pathology - Bogotá D. C. - Colombia.
6 Universidad Nacional de Colombia - Bogotá Campus - Institute of Genetics - Bogotá D. C. - Colombia
.Open access
Received: 22/07/2021
Accepted: 28/03/2022
Corresponding author: Diana Carolina Grajales-Ospina. Grupo de Investigación Patología Molecular, Facultad de Medicina, Universidad Nacional de Colombia. Bogotá D.C. Colombia. Email: dgrajaleso@unal.edu.co.
Keywords: Noninvasive Prenatal Testing; Aneuploidy; High-Risk Pregnancy; Karyotype (MeSH).
Palabras clave: Prueba prenatal no invasiva; Aneuploidia, Embarazo de alto riesgo; Cariotipo (DeCS).
How to cite: Grajales-Ospina DC, Yunis-Hazbun LK, Camacho-Montaño AM, Bautista-Charry AA, Niño-Alba R, Yunis JJ. Concordance analysis between noninvasive prenatal testing (NIPT) and prenatal karyotyping for detecting fetal aneuploidies. Rev. Fac. Med. 2023;71(2):e97438. English. doi: https://doi.org/10.15446/revfacmed.v71n2.97438.
Cómo citar: Grajales-Ospina DC, Yunis-Hazbun LK, Camacho-Montaño AM, Bautista-Charry AA, Niño-Alba R, Yunis JJ. [Análisis de concordancia entre la prueba prenatal no invasiva (NIPT) y el cariotipo prenatal para la detección de aneuploidías fetales]. Rev. Fac. Med. 2023;71(2):e97438. English. doi: https://doi.org/10.15446/revfacmed.v71n2.97438.
Copyright: Copyright: ©2023 Universidad Nacional de Colombia. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, as long as the original author and source are credited.
Abstract
Introduction: Noninvasive prenatal testing (NIPT) is a screening test for fetal aneuploidy with higher specificity and sensibility rates compared to traditional biochemical prenatal screening.
Objective: To evaluate concordance between NIPT and prenatal karyotyping testing for detecting fetal aneuploidies in pregnancies with high risk of such disorders.
Materials and methods: Prospective pilot study conducted between September 2019 and December 2020 in 20 pregnant patients classified as high risk of aneuploidy based on ultrasound findings and treated in Bogotá and
Medellín, Colombia. Each patient underwent a confirmatory invasive test (karyotyping) and a NIPT and an invasive confirmatory test (prenatal karyotyping). Concordance between both methods was determined using Cohen's kappa coefficient (significance level p<0.05), where values >0.7 were considered as a good level of agreement.
Results: Aneuploidies were detected in 3 of the 20 pregnancies (15%) by means of invasive cytogenetic testing, namely, trisomy 21, trisomy 18, and monosomy X. NIPT detected the trisomy 21 and monosomy X cases but failed to detect trisomy 18. Regarding the concordance analysis between NIPT and prenatal karyotype testing in the detection of aneuploidies, Cohen's kappa coefficient was 0.77. As for their concordance for the detection of trisomy 21 and X monosomy, Cohen's kappa coefficient was 1.0, while it was 0 for the detection of trisomy 18. In addition, NIPT detected 67% of aneuploidies.
Conclusion: The results of this study, the first of its kind to be conducted in Colombia, showed good concordance between NIPT and prenatal invasive testing (karyotyping) for detecting fetal aneuploidies. However, the results obtained stress the recommendation of using NIPT only as a screening test and not as a diagnostic test.
Resumen
Introducción. La prueba prenatal no invasiva (NIPT, por su sigla en inglés) es una prueba de tamización de aneuploidías fetales con una mayor sensibilidad y especificidad que la tamización bioquímica prenatal tradicional.
Objetivo. Evaluar la concordancia entre la NIPT y el cariotipo prenatal para la detección de aneuploidías fetales en embarazos de alto riesgo de dichas anomalías.
Materiales y métodos. Estudio piloto prospectivo realizado entre septiembre de 2019 y diciembre de 2020 en 20 pacientes con gestaciones clasificadas como de alto riesgo para aneuploidías fetales con base en los hallazgos ecográficos y atendidas en Bogotá y Medellín, Colombia. A cada paciente se le realizó una NIPT y una prueba invasiva confirmatoria (cariotipo prenatal). La concordancia entre ambos métodos se determinó mediante el coeficiente kappa de Cohen (nivel de significancia p<0.05), donde valores >0.7 se consideraron como un buen nivel de concordancia.
Resultados. En 3 de las 20 gestaciones (15%) se detectaron aneuploidías mediante estudio citogenético invasivo: trisomía 21, trisomía 18 y monosomía X. La NIPT detectó la trisomía 21 y la monosomía X, pero falló en detectar la trisomía 18. En lo que respecta a la concordancia entre la NIPT y el cariotipo prenatal para la detección de aneuploidías, el coeficiente de kappa de Cohen fue 0.77; en el caso de su concordancia para la detección de la trisomía 21 y la monosomía X el coeficiente de kappa de Cohen fue 1, mientras que para la detección de la trisomía 18 fue 0. Además, la NIPT detectó 67% de las aneuploidías.
Conclusión. En el presente estudio, primero en realizarse en Colombia, se observó una buena concordancia entre la NIPT y la prueba invasiva (cariotipo prenatal) para la detección de aneuploidías. Sin embargo, los resultados aquí reportados enfatizan la recomendación de utilizar la NIPT como prueba de tamización y no como prueba diagnóstica.
Introduction
In Colombia, according to a study conducted by Zarante et al.1 in which 52 744 births registered between 2001 and 2008 in Bogotá, Ubaté and Manizales were analyzed, 3.122% of newborns had some type of congenital malformation. Congenital structural malformations account for about 50% of congenital diseases, of which approximately 24% are due to copy number variations, 15% to single gene disorders, and a little more than 10% to chromosomal abnormalities; of the latter, 80-85% are common trisomies (trisomy 13, 18, and 21) and sex chromosome aneuploidies.2
Aneuploidies are one of the main causes of perinatal death and cognitive disability,3,4 and this has led to the development of prenatal screening programs that take into account ultrasound findings and serum test results, which, together with maternal age, allow suspecting the presence of chromosomal alterations in the fetus.3,5,6 Detection rates for this type of alteration vary depending on the trimester of the pregnancy; for example, in the first trimester, they are 85-90% and in the second trimester, 61-70%.5,7 In Colombia, according to the Ministry of Health, neonatal screening should be based on maternal age and nuchal translucency measurement.8 If this screening is found to be altered, invasive cytogenetic testing should be performed.9
Since 2011, noninvasive prenatal testing (NIPT) has been available on the market as a screening test for the detection of fetal aneuploidies, mainly in chromosomes 13, 18 and 21, and in sex chromosomes,10 and in recent years, this test has also been offered by several commercial companies for the additional detection of microdeletions/microduplications.10 NIPT entails the detection of cell-free fetal DNA in maternal plasma,11 which originates from trophoblast apoptosis,2,12 and is performed by massively parallel sequencing and advanced bioinformatics analysis.7,11 “Fetal fraction”, which is the percentage of total cell-free fetal DNA circulating in maternal plasma, is a major factor for the performance of this test since at least 4% fetal fraction is required for obtaining an accurate result.2,12 There are several biological factors that influence fetal fraction, such as gestational age, body mass index, type of aneuploidy, ethnicity, chromosomal mosaicism, assisted fertilization, use of heparins during pregnancy, vanishing twin, and neoplasms detected during gestation.2,10-13
NIPT is a reliable method that has lower false positive rates than traditional prenatal screening for the detection of fetal aneuploidy.12 In a systematic review and meta-analysis published 2016, Taylor-Phillips et al.14 found that the sensitivity of NIPT for trisomy 21, 18, and 13 was 99.3% (95%CI: 98.9-99.6), 97.4% (95%CI: 95.8-98.4), and 97.4% (95%CI: 86.1-99.6),
respectively. A year later, Skrzypek & Hui7 found that the overall false positive rate of NIPT for common aneuploidies and sex chromosomes was 0.72%. However, the clinical usefulness of this test for detecting microdeletions/microduplications is still unclear, as its effectiveness rate varies in the different studies performed.15,16
It should be kept in mind that although NIPT performs well in detecting common fetal aneuploidies, it is currently considered a screening test7 and the gold standard for detecting these anomalies is prenatal karyotyping,9 even though it is an invasive technique.
In view of the above, the objective of the present study was to evaluate concordance between NIPT and prenatal karyotyping for the detection of fetal aneuploidies in pregnancies at high risk of such disorders.
Materials and methods
Study type and population
Prospective case series study conducted between September 2019 and December 2020. Women under 22 weeks gestation whose pregnancies were classified as high risk for aneuploidy based on ultrasound findings and, therefore, required confirmatory invasive testing were included.
Convenience sampling was used to obtain the sample, considering the following exclusion criteria: multiple gestation, presence of neoplasms in the mother, history of transplantation, and history of heparin use or use during pregnancy. Thus, 21 pregnant women were included: 20 treated at the Instituto Materno Infantil in Bogotá (Colombia) and 1 treated in a private practice in Medellín (Colombia). However, one of the cases did not complete all the studies because, although the initial ultrasound (performed in week 14) diagnosed a possible omphalocele, on the day scheduled for the invasive test (4 weeks later) this defect was not found, only an umbilical cyst, for which reason a confirmatory chromosomal study (prenatal karyotype) was not performed since it was considered that, based on this ultrasound finding, the pregnancy was not at high risk for aneuploidy. Thus, the final sample consisted of 20 pregnant women.
Procedures
Data collection
Clinical and sociodemographic information on each patient was collected by means of an interview conducted on the day the invasive test was performed or at a consultation prior to the procedure. On the day of the test, peripheral blood samples were also collected for NIPT (all patients) and for biochemical and hormonal tests used for prenatal screening (15 patients).
Sample collection
Noninvasive testing
Serum tests: plasma serum screening studies were performed in plasma during the first trimester (pregnancy-associated plasma protein A and β-human chorionic gonadotropin) or second trimester (β-human chorionic gonadotropin, alpha-fetoprotein, and unconjugated estriol) by venipuncture. Samples were sent to a reference laboratory for processing but as mentioned above, these studies were performed in only 15 patients for comparative purposes.
NIPT test: blood samples were collected in Cell-Free DNA BCT and EDTA tubes between 13 and 22 weeks based on ultrasound diagnosis. Plasma separation was done within the first 6 hours and kept frozen at -80°C. For the isolation of circulating fetal DNA in maternal blood, the QIAamp Circulating Nucleic Acid (QIAGEN®) kit was used following the manufacturer’s recommendations. Circulating fetal DNA was stored at -80°C until processing. Once there were 6 samples for each assembly, genetic libraries were prepared to perform next generation sequencing as instructed by the manufacturer of the NIPTSG-BabyTest kit; this test detects fetal aneuploidies for chromosomes X, Y, 21, 13 and 18, as well as various copy number variations (microdeletion/microduplication). Libraries were analyzed on the Illumina MySeq platform using standard 300-cycle MySeq cells and following the NIPT kit manufacturer’s recommendations. FastQ files obtained from the sequencing were analyzed on the GeneSystems Platform.
Invasive testing (prenatal karyotyping)
An amniotic fluid sample was collected from 19 patients after week 16 of pregnancy, and one patient underwent chorionic villus biopsy at week 10, obtaining between 10mL and 20mL of amniotic fluid for each sample (samples and biopsy were taken by the attending obstetrician in all cases). A conventional chromosomal study was performed on all samples and once the prenatal karyotype result was obtained, it was reported to the treating physician of each patient.
For invasive test processing, the amniotic fluid was centrifuged, and the cell culture was incubated in duplicate plates in special culture media for amniocyte growth. For the case of chorionic villi, the sample was manually disaggregated and then centrifuged; the cell culture was incubated in duplicate plates in a special culture media for trophoblast cell growth. After 7 to 12 days, standard treatments were performed to visualize the chromosomes as per institutional protocols, and cytogenetic analysis was performed using the Case Data Manager system of Applied Spectral Imaging V.8.0.
Statistical analysis
The sociodemographic and clinical data of the 20 patients were collected in a Microsoft Excel spreadsheet. Data were analyzed by means of descriptive statistics using absolute frequencies and percentages, as well as measures of central tendency (mean) and dispersion (minimum and maximum). Moreover, Cohen’s kappa coefficient was used to determine the concordance between both methods (NIPT and prenatal karyotyping) for the detection of aneuploidies, the detection of each of the cytogenetic alterations identified, and the determination of fetal sex (significance level p<0.05), where values >0.7 were considered as a good level of concordance.
Ethical considerations
The study was approved by the Ethics Committee of the Faculty of Medicine of the Universidad Nacional de Colombia, as stated in Minutes No. 015-179 of August 15, 2019. The ethical principles for research involving human subjects established in the Declaration of Helsinki17 and the health research provisions contained in Resolution 8430 of 1993 of the Colombian Ministry of Health were also taken into account.18 Prior to their participation in the study, all patients signed an informed consent form.
Results
The mean age of the participants was 30 years (range 17-42) and most (65%) were older than 35 years. Samples for NIPT were taken on average at week 17 of pregnancy (range 13-22 weeks). The clinical and sociodemographic characteristics of the patients are shown in Table 1.
Table 1. Clinical and sociodemographic characteristics of the pregnant women included in the study (n=20).
|
Characteristic
|
n (%)
|
|
Age (mean, range)
|
30 years old (17-42)
|
|
Gestational age (mean, range)
|
17 weeks (13-22)
|
|
Body mass index
|
Normal
|
14 (70%)
|
|
Overweight
|
4 (20%)
|
|
Obesity
|
2 (10%)
|
|
Parity
|
Nulliparous
|
4 (20%)
|
|
Multiparous
|
16 (80%)
|
|
Socioeconomic level
|
Low
|
19 (95%)
|
|
Medium
|
1 (5%)
|
|
Smoking
|
No
|
20 (100%)
|
|
Diabetes mellitus
|
No
|
20 (100%)
|
|
History of pregnancy with trisomy 21
|
No
|
19 (95%)
|
|
Yes
|
1 (5%)
|
Source: Own elaboration.
Table 2 presents ultrasound findings, NIPT results, prenatal karyotype results, and the therapeutic approach adopted for each case. As mentioned above, one of the pregnant women initially included (patient 4) was excluded because she was diagnosed with omphalocele in the first ultrasound evaluation (week 14), but at the time of the invasive test (4 weeks later) this defect was not found, but rather a cyst of the umbilical cord, so the pregnancy was not considered to be at high risk of aneuploidy. However, the data of this patient are included in Table 2 for descriptive purposes.
Table 2. Ultrasound findings, cytogenetic results, noninvasive prenatal test results, and gestational therapeutic approach and follow-up.
|
Patient
|
Ultrasound findings
|
Prenatal cytogenetics
|
Noninvasive prenatal test result
|
Therapeutic approach to pregnancy
|
|
1
|
Nasal hypoplasia, increased NT
|
47,XX,+21
|
High risk for T21
|
VTP
|
|
2
|
Increased NT
|
46,XY
|
Low risk
|
Delivery of a healthy baby
|
|
3
|
Increased NT
|
46,XX
|
Low risk
|
Delivery of a healthy baby
|
|
4 *
|
Umbilical cord cyst
|
Not performed
|
Low risk
|
Delivery of a healthy baby
|
|
5
|
Cystic hygroma, complex heart disease, hydrops fetalis, left hydronephrosis, single umbilical artery
|
45,X
|
High risk for monosomy X
|
VTP
|
|
6
|
Increased NT
|
46,XY
|
Low risk
|
Delivery of a healthy baby
|
|
7
|
Central nervous system with severe ventriculomegaly, atrioventricular canal defect, severe IUGR
|
46,XY
|
Low risk
|
VTP
|
|
8
|
Sinus bradyarrhythmia, VSD, right atrial enlargement, heterotopic gastric mucosa
|
46,XX
|
Low fetal DNA fraction
|
Cesarean section, infant with heart disease under study, and heterotopic gastric mucosa
|
|
9
|
Increased NT
|
46,XX
|
Low risk
|
VTP
|
|
10
|
Urinary tract obstruction, increased bladder size
|
46,XY
|
Low risk
|
VTP
|
|
11
|
Atrioventricular canal defect and micrognathia
|
46,XY
|
Low risk
|
Delivery of a healthy baby
|
|
12
|
Severe hydrocephalus, occipital interhemispheric cyst
|
46,XY
|
Low risk
|
VTP
|
|
13
|
Increased NT
|
46,XY
|
Low risk
|
Delivery of a healthy baby
|
|
14
|
Bilateral choroid plexus cyst, microcephaly, VSD
|
47,XY,+18
|
Not informative for T18
|
VTP
|
|
15
|
IUGR, left choroidal cyst, severe megacystic bladder, bilateral renal dysplasia
|
46,XY
|
Low risk
|
VTP
|
|
16
|
Increased NT
|
46,XX
|
Low risk
|
Delivery of a healthy baby
|
|
17
|
Increased NT
|
46,XY
|
Low risk
|
Delivery of a healthy baby
|
|
18
|
Increased NT
|
46,XX
|
Low risk
|
Delivery of a healthy baby
|
|
19
|
Megabladder, single umbilical artery
|
46,XX
|
Low risk
|
Baby with anal atresia under follow-up
|
|
20
|
VSD
|
46,XY
|
Low risk
|
Delivery of a healthy baby
|
|
21
|
Increased NT
|
46,XX
|
Low risk
|
Delivery of a healthy baby
|
NT: nuchal translucency; VTP: voluntary termination of pregnancy; T21: trisomy 21; IUGR: intrauterine growth restriction; VSD: ventricular septal defect.
* Case excluded from analysis.
Source: Own elaboration.
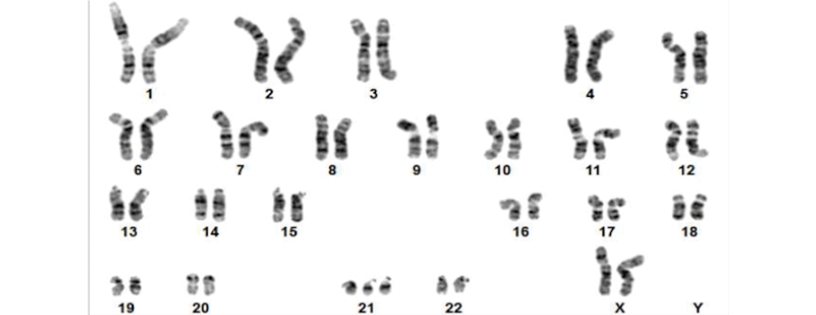
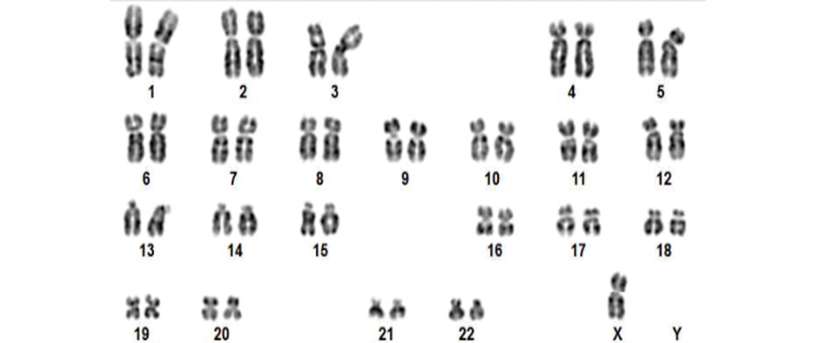
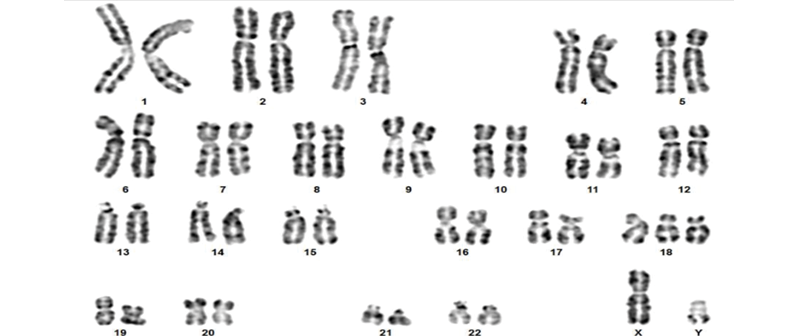
Of the 20 cases analyzed, aneuploidies were detected by prenatal karyotyping in only 3 cases (15%): a trisomy 21 (Figures 1 and 2), a monosomy X (Figures 3 and 4), and a trisomy 18 (Figures 5 and 6).
Figure 1. Patient 1. Karyotype with G-banding, 47,XX,+21, typical of Down syndrome.
Source: Image obtained while conducting the study.
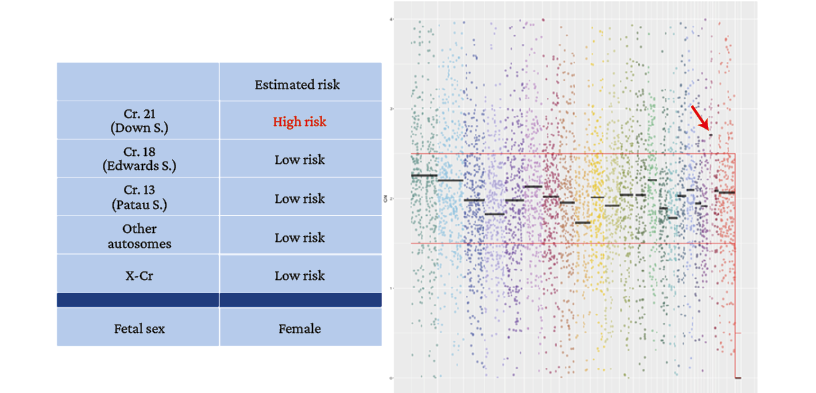
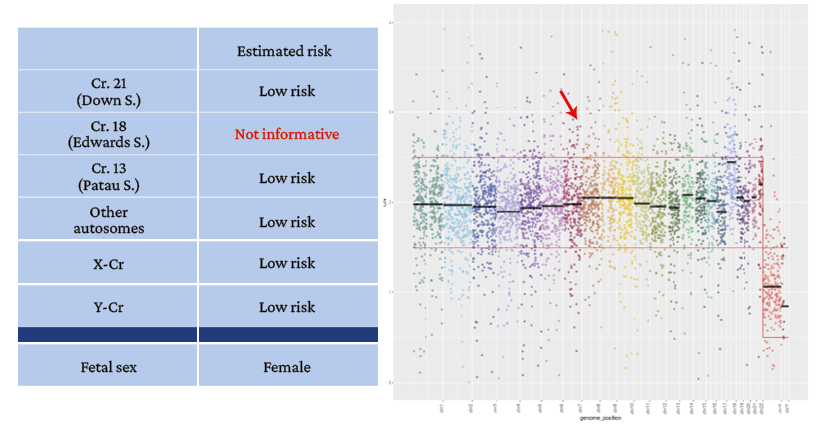
Figure 2. Patient 1. Noninvasive prenatal test result. Female fetal sex.
→ Gain of chromosome 21 above the average calculated as normal (high risk for trisomy 21).
Source: Image obtained while conducting the study.
Figure 3. Patient 5. Karyotype with G-banding, 45,X indicating female fetal sex with monosomy X, typical of Turner syndrome.
Source: Image obtained while conducting the study.
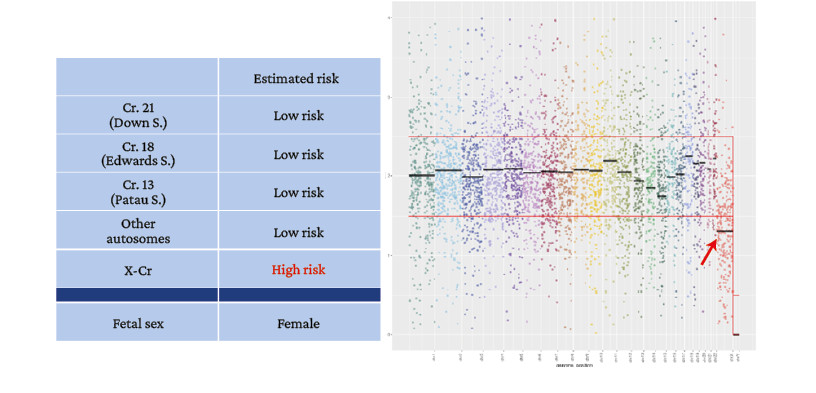
Figure 4. Patient 5. Noninvasive prenatal test result. Female fetal sex due to absence of the Y chromosome.
→ Loss of genetic information on the X chromosome below the average expected to classify test as normal (high risk for monosomy X).
Source: Image obtained while conducting the study.
Figure 5. Patient 14. Karyotype with G-banding, 47,XY,+18 indicating male fetal sex with trisomy 18, typical of Edwards syndrome.
Source: Image obtained while conducting the study.
Figure 6. Patient 14. Noninvasive prenatal test result. Male fetal sex.
→ Gain of chromosome 18 at the upper normal limit, without exceeding the average calculated as normal.
Source: Image obtained while conducting the study.
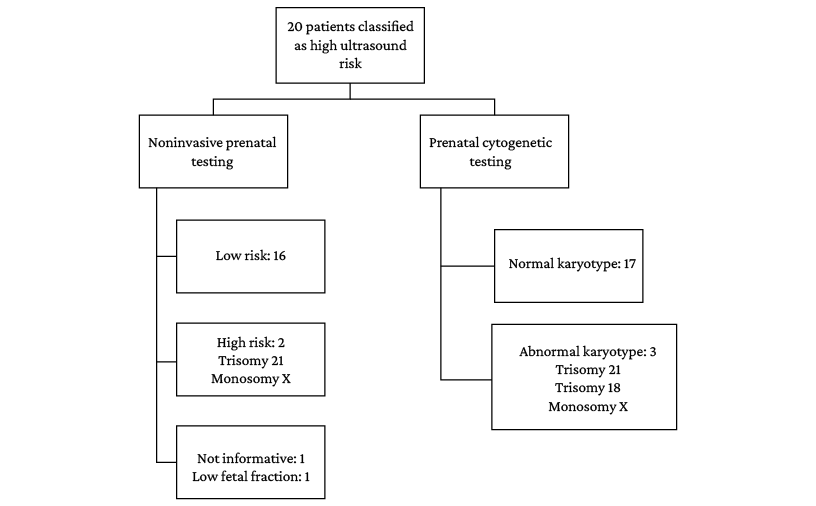
Despite repeating the isolation of the fetal DNA fraction in maternal blood on two occasions, one of the samples could not be analyzed for low fetal DNA fraction. In the remaining samples, the NIPT yielded the following results: 1 high-risk case for trisomy 21 (Figure 2), 1 high-risk case for monosomy X (Figure 4), 16 low-risk cases of aneuploidy, and 1 case classified as inconclusive for trisomy 18 on the GenSystems Platform bioinformatics platform (Figure 6). In the latter case, the test could not be repeated as the parents decided to voluntarily terminate the pregnancy based on the cytogenetic findings.
No cases with microdeletion/microduplication were detected in the NIPT. Thus, the test used detected aneuploidies in 67% of the cases of aneuploidies detected by cytogenetic study. The comparison of the findings obtained in each type of test is presented in Figure 7.
Figure 7. Results of confirmatory invasive testing (prenatal karyotyping) and noninvasive prenatal testing.
Source: Own elaboration.
Concordance between NIPT and prenatal karyotyping
Regarding overall concordance between NIPT and prenatal karyotyping for the detection of aneuploidy, a Cohen’s kappa coefficient of 0.77 was obtained, indicating good concordance, which was also statistically significant (p=0.0006). When discriminating for trisomy, Cohen’s kappa coefficient was 1 for both trisomy 21 (p=0.0001) and monosomy X (p=0.0001) detection, representing excellent concordance (Table 3); however, concordance for trisomy 18 was 0%. Concerning fetal sex determination, the Kappa index was 1 (p=0.0001), i.e., 100% concordance.
Table 3. Concordance between noninvasive prenatal testing and prenatal karyotyping.
|
Fetal aneuploidy screening
|
Trisomy 21 screening
|
Trisomy 18 screening*
|
Monosomy X screening
|
Fetal sex screening
|
|
Kappa coefficient
|
0.77
(p=0.0006)
|
1
(p=0.0001)
|
0
|
1
(p=0.0001)
|
1
(p=0.0001)
|
* Failure to detect trisomy 18 by noninvasive prenatal testing.
Source: Own elaboration.
Combined screening
The standard biochemical screening performed in the first trimester was carried out in only 8 of 9 pregnant women in this period of pregnancy. All these pregnancies were classified as high risk by serum screening, whereas only 25% were classified as high risk by NIPT. When performing the confirmatory test, NIPT correctly detected the two pregnancies at high risk of fetal aneuploidies (trisomy 21 and monosomy X).
When screening during the second trimester, serum screening was performed in only 7 of 11 patients who were in this period of gestation. Of these 7, 4 (57.14%) were classified as high risk for aneuploidies, 1 (14.28%) as intermediate risk, and 2 (28.57%) as low risk. When NIPT was performed, all were classified as low risk, with the exception of the case with trisomy 18, whose result was inconclusive (not informative). Although the number of samples analyzed was low, it was evident that there is a higher overestimation of aneuploidy risk in the second trimester when using serum tests compared to NIPT (71.42% vs. 14.28%). Serum screening tests showed high risk of aneuploidy in 80% of cases (12/15), moderate risk in 6.66% (1/15), and low risk in 13.33% (2/15). Overall, 86.66% of the 15 patients who underwent these tests were classified as being at increased risk of aneuploidy.
Postnatal follow-up
40% (n=8) of the pregnancies analyzed in the present study were terminated by the parents based on ultrasound and cytogenetic findings. Of the pregnancies that made it to term (n=12), it was established that 10 babies were born healthy and 2 required medical follow-up, one (patient 8) due to gastric ectopia and congenital heart disease, and another (patient 19) due to anal atresia.
Discussion
The present study found that while prenatal karyotyping detected aneuploidies in 3 pregnant women (trisomy 21, trisomy 18, and monosomy X), only two were detected with NIPT (trisomy 21 and monosomy X), failing to detect trisomy 18. This can be explained because the signal for chromosome 18 in NIPT was found at the upper limit without exceeding the abnormality threshold, so the bioinformatics algorithm did not detect this disorder.
The overall concordance between both tests, measured with Cohen’s kappa coefficient, was 0.77 with a p=0.0006, indicating good concordance. Moreover, when concordance analysis was performed for each detected trisomy, a concordance of 1 was obtained for trisomy 21 and monosomy X, but 0 for trisomy 18. In this regard, Guy et al.,19 in a study of 69 749 samples from women with singleton pregnancies, found that the overall sensitivity of NIPT for detecting major aneuploidies was 97.9%, with a specificity of 99.9% and a positive predictive value (PPV) of 87.2%. These authors also found that the PPV for trisomy 21, 18, and 13 was 98.1%, 88.2%, and 59.3%, respectively, and for sex chromosome aneuploidies and microdeletions was 69% and 75%, respectively.19
In a previous meta-analysis including 35 studies, Gil et al.20 found that the NIPT detection rate for trisomy 21 was 99.7% (95%CI: 99.1-99.9), with a false-positive rate of 0.04% (95%CI: 0.02-0.07);
that it was 97.9% (95%CI: 94.9-99.1) for trisomy 18, with a false-positive rate of 0.04% (95%CI: 0.03-0.07); that it was 99.0% (95%CI: 65.8-100) for trisomy 13, with a false positive rate of 0.04% (95%CI: 0.02-0.07); and that it was 95.8% (95%CI: 70.3-99.5) for monosomy X, with a false positive rate of 0.14% (95%CI: 0.05-0.38). It can be seen that NIPT sensitivity for trisomy 18 in the Gil et al.20 meta-analysis showed that up to 2.1% of cases of this disorder yielded a false-
negative result, as was the case in the present study.
The screening performance of NIPT depends on several factors. Gil et al.,20 for example, found three reasons for the low performance of this test in the detection of aneuploidies: 1) issues with sample collection and transportation (inadequate sample volume, hemolysis, or misidentification errors), 2) low fetal fraction, usually <4%, and 3) sample processing failures during DNA extraction, amplification, or sequencing. The main reason for a failed result is a low fetal fraction, which can be explained by maternal obesity and low placental size. In trisomies 18 and 13, fetal fraction is usually lower than in trisomy 21, so it can be considered that there is a higher risk for test failure in trisomies 18 and 13.13
In case of unsuccessful results due to low fetal DNA fraction using NIPT, it is recommended to take a new sample, since fetal fraction increases as gestational age increases. However, performing this procedure would delay the detection of fetal aneuploidies and thus reduce the ability to define whether additional testing is necessary,21 as was the case of patient 8 in the present study, who ultimately could not be included in the concordance analysis.
As reported by the Committee on Genetics Society for Maternal-Fetal Medicine, NIPT has a failure rate ranging from 1% to 8%, which varies depending on the laboratory and the methodology used.12 In turn, Gil et al.20 report non-detection rates of 5.9% for trisomies in autosomes and 11.7% for aneuploidies in sex chromosomes. Also, as stated by Van-Opstal et al.,22 there are other biological factors that can affect the detection rate of NIPT, such as the origin of the fetal DNA (cytotrophoblast) and chromosomal mosaicism (<30% is not detected by NIPT), so it is not possible to achieve 100% sensitivity and specificity. These authors further reported that the estimated probability of a false negative NIPT varies between 0.02 and 0.26%.22
In the present study, the false negative rate was 5.26% (1/19 cases); however, these data should be treated with caution because of the sample size. In the case of patient number 14, for example, the cytogenetics result showed universal trisomy 18 and her fetal DNA fraction was >4% (9.4%), demonstrating the importance of interpreting NIPT results together with a medical geneticist who is knowledgeable about the biological limitations of the test in order to provide better advice to the patient.
According to the Colombian Ministry of Health,8 screening for fetal malformations should be based on maternal age and ultrasound measurement of nuchal translucency.
In a study conducted in Cali (Colombia), Fandiño-Losada et al.23 found that of 738 karyotype records analyzed, 14% had chromosomal alterations and that the most frequent indications for an invasive procedure for prenatal diagnosis were single anatomical alteration in second trimester ultrasound (21.4%) and maternal age (18.8%). In the present study, 12 of the 20 patients were screened with biochemical tests to cross-check the NIPT results. For first trimester screening, with all cases being high risk according to ultrasound, 100% of the pregnant women were classified as high risk in biochemical screening, but only 25% showed high risk in NIPT. For second trimester screening, 71.42% of pregnancies were classified as high risk by biochemical screening, while NIPT showed low risk in all cases, except for the pregnancy in which trisomy 18 was diagnosed, as NIPT yielded an uninformative or inconclusive result.
The results of the present study emphasize the recommendation to use NIPT only as a screening test due to possible false negatives (trisomy 18 in this study) or false positives, as described in the literature,20,22 and not as a diagnostic test, even more so in pregnancies at high risk of aneuploidy. Likewise, the results demonstrate the need to establish multidisciplinary work teams when using NIPT, involving geneticists to correlate noninvasive and invasive test findings with ultrasound findings.
A limitation of the present study was the low number of cases analyzed, so it was not possible to estimate the sensitivity, specificity, and predictive values of the NIPT used. However, the results reported here demonstrate that NIPT can be carried out in Colombia without the need to send samples abroad, as is currently the case.
Conclusions
In the present study, the first of its kind in Colombia that compares NIPT with prenatal karyotyping for the detection of aneuploidies in high-risk pregnancies for these anomalies, there was good concordance between the two methods (67%, Kappa index = 0.77). However, the results emphasize the recommendation to use NIPT as a screening test and not as a diagnostic test.
Conflicts of interest
None stated by the authors.
Funding
This project was funded by the Universidad Nacional de Colombia through the research project “Análisis de concordancia de pruebas no invasivas prenatales de sangre materna (NIPT) con prueba invasiva para embarazos de alto riesgo de aneuploidías fetales en una muestra piloto” (code HERMES 47848) and by the IPS Servicios Médicos Yunis Turbay y Cia. SAS, where the samples were processed for the confirmatory cytogenetic studies and NIPT by next generation sequencing (NGS).
Acknowledgments
To the patients who voluntarily agreed to participate in this study, and to the Subred Integrada de Servicios de Salud Centro Oriente E.S.E., for allowing the recruitment of patients at the Instituto Materno Infantil de Bogotá.
References
1.Zarante I, Franco L, López C, Fernández N. Frecuencia de malformaciones congénitas: evaluación y pronóstico de 52.744 nacimientos en tres ciudades colombianas. Biomédica. 2010;30(1):65-71. https://doi.org/b4sm.
2.Shaffer BL, Norton ME. Cell-Free DNA Screening for Aneuploidy and Microdeletion Syndromes. Obstet Gynecol Clin North Am. 2018;45(1):13-26. https://doi.org/gc8397.
3.Carlson LM, Vora NL. Prenatal Diagnosis: Screening and Diagnostic Tools. Obstet Gynecol Clin North Am. 2017;44(2):245-56. https://doi.org/f99h39.
4.Nussbaum RL, McInnes RR, Willard HF. Thompson & Thompson Genetics in Medicine E-Book. Philadelphia: Elsevier Health Sciences; 2015.
5.Practice Bulletin No. 163: Screening for Fetal Aneuploidy. Obstet Gynecol. 2016;127(5):e123-37. https://doi.org/gmmb3q.
6.Norwitz ER, Levy B. Noninvasive prenatal testing: the future is now. Rev Obstet Gynecol. 2013;6(2):48-62.
7.Skrzypek H, Hui L. Noninvasive prenatal testing for fetal aneuploidy and single gene disorders. Best Pract Res Clin Obstet Gynaecol. 2017;42:26-38. https://doi.org/gbrdww.
8.Colombia. Ministerio de Salud y Protección Social (MinSalud) - Colciencias. Guía de Práctica Clínica para la prevención, detección temprana y tratamiento del embarazo, parto o puerperio. Guía No. 11-15. Bogotá D.C.:
MinSalud; 2013.
9.Costa JM, Letourneau A, Favre R, Bidat L, Belaisch-Allart J, Jouannic JM, et al. Cell-free fetal DNA versus maternal serum screening for trisomy 21 in pregnant women with and without assisted reproduction technology: a prospective interventional study. Genet Med. 2018;20(11):1346-53. https://doi.org/kc4g.
10.Allyse M, Minear MA, Berson E, Sridhar S, Rote M, Hung A, et al. Non-invasive prenatal testing: a review of international implementation and challenges. Int J Womens Health. 2015;7:113-26. https://doi.org/ghvmzg.
11.Audibert F, De Bie I, Johnson JA, Okun N, Wilson RD, Armour C, et al. No. 348-Joint SOGC-CCMG Guideline: Update on Prenatal Screening for Fetal Aneuploidy, Fetal Anomalies, and Adverse Pregnancy Outcomes. J Obstet Gynaecol Can. 2017;39(9):805-17. https://doi.org/kdf3.
12.Committee Opinion No. 640: Cell-Free DNA Screening For Fetal Aneuploidy. Obstet Gynecol. 2015;126(3):e31-7. https://doi.org/ghvmrk.
13.Revello R, Sarno L, Ispas A, Akolekar R, Nicolaides KH. Screening for trisomies by cell-free DNA testing of maternal blood: consequences of a failed result. Ultrasound Obstet Gynecol. 2016;47(6):698-704. https://doi.org/f9pr8m.
14.Taylor-Phillips S, Freeman K, Geppert J, Agbebiyi A, Uthman OA, Madan J, et al. Accuracy of non-invasive prenatal testing using cell-free DNA for detection of Down, Edwards and Patau syndromes: a systematic review and meta-analysis. BMJ Open. 2016;6(1):e010002. https://doi.org/f8kbxd.
15.Wapner RJ, Martin CL, Levy B, Ballif BC, Eng CM, Zachary JM, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367(23):2175-84. https://doi.org/ggh62d.
16.Petersen AK, Cheung SW, Smith JL, Bi W, Ward PA, Peacock S, et al. Positive predictive value estimates for cell-free noninvasive prenatal screening from data of a large referral genetic diagnostic laboratory.
Am J Obstet Gynecol. 2017;217(6):691.e1-6. https://doi.org/ghwgt2.
17.World Medical Association (WMA). WMA Declaration of Helsinki – Ethical principles for medical research involving human subjects. Fortaleza: 64th WMA General Assembly; 2013.
18.Colombia. Ministerio de Salud. Resolución 8430 de 1993 (octubre 4): Por la cual se establecen las normas científicas, técnicas y administrativas para la investigación en salud. Bogotá D.C.; october 4 1993.
19.Guy C, Haji-Sheikhi F, Rowland CM, Anderson B, Owen R, Lacbawan FL, et al. Prenatal cell-free DNA screening for fetal aneuploidy in pregnant women at average or high risk: Results from a large US clinical laboratory. Mol Genet Genomic Med. 2019;7(3):e545. https://doi.org/kdf5.
20.Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. 2017;50(3):302-14. https://doi.org/gm5pgt.
21.Luo Y, Hu H, Zhang R, Pan Y, Ma Y, Long Y, et al. [Factors affecting the failure of non-invasive prenatal testing and the feasibility analysis of retesting]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2020;37(6):603-8.
22.Van-Opstal D, Srebniak MI, Polak J, de Vries F, Govaerts LC, Joosten M, et al. False Negative NIPT Results: Risk Figures for Chromosomes 13, 18 and 21 Based on Chorionic Villi Results in 5967 Cases and Literature Review. PLoS One. 2016;11(1):e0146794. https://doi.org/f8mmxw.
23.Fandiño-Losada A, Lucumí-Villegas B, Ramírez-Cheyne J, Isaza-de, Lourido C, Saldarriaga C. Valor predictivo positivo del diagnóstico prenatal invasivo para alteraciones cromosómicas. Rev. Fac. Med. 2018;66(1):19-24. https://doi.org/kdf7.
 Luz Karime Yunis-Hazbun1,2
Luz Karime Yunis-Hazbun1,2 Andrés Mauricio Camacho-Montaño3
Andrés Mauricio Camacho-Montaño3 Alejandro Antonio Bautista-Charry4
Alejandro Antonio Bautista-Charry4 Reinaldo Niño-Alba4
Reinaldo Niño-Alba4 Juan José Yunis1,2,5,6
Juan José Yunis1,2,5,6