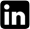POPULATION GENETICS OF Atta sexdens rubropilosa (HYMENOPTERA: FORMICIDAE)
The genetic variability of Atta sexdens rubropilosa leaf-cutting ants collected from five brazilian localities was evaluated with PCR-RAPD technique. We used 15 primers producing 148 fragments of which 123 (83,11 %) contained polymorphisms. The estimated Shannon index was 0.3836 ± 0.2335 showing that these ants possess high genetic diversity. The GST value was 0,2372 and PT = 0,184, indicating that the analyzed populations are moderately differentiated and 82 % of the variation obtained occur within populations. Although Mantel’s test had shown correlation between genetic distances and geographic was observed that Ivatuba and Itambé (33,8 km) have the small geographical distance and the largest genetic distance. The lower genetic distance was estimated for Maringá and Ivatuba but this localities have a small geographic distance (42,3 km), indicating that there are no barriers for mating among reproducers in these populations. The high degree of polymorphism (83,11 %) and the ability to cross among the populations in the studied regions indicate that this species of leaf-cutting ant is well adapted to the region; therefore, integrated control programs can be developed.
POPULATION GENETICS OF Atta sexdens rubropilosa (HYMENOPTERA: FORMICIDAE)
Genética de poplaciones de Atta sexdens rubropilosa (Hymenoptera: Formicidae)
LIRIANA BELIZÁRIO CANTAGALLI1, Ph. D.; CLAUDETE APARECIDA MANGOLIN2, Ph. D.; MARIA CLAUDIA COLLA RUVOLO TAKASUSUKI2, Ph. D.
1 Departamento de Biotecnología, Genética y Biología Celular, Universidad Estatal de Maringá. 2 Docente Departamento de Biotecnología, Genética y Biología Celular, Universidad Estatal de Maringá. Av. Colombo, 5790, CEP: 87020900, Maringá, Paraná, Brasil. Tel.: 44 3011 4681 Corresponding author: mccrtakasusuki@uem.br
Presentado el 25 de mayo de 2012, aceptado el 27 de septiembre de 2012, correcciones el 7 de febrero de 2013.
ABSTRACT
The genetic variability of Atta sexdens rubropilosa leaf-cutting ants collected from five brazilian localities was evaluated with PCR-RAPD technique. We used 15 primers producing 148 fragments of which 123 (83.11 %) contained polymorphisms. The estimated Shannon index was 0.3836 ± 0.2335 showing that these ants possess high genetic diversity. The GST value was 0.2372 and ΦPT = 0.184, indicating that the analyzed populations are moderately differentiated and 82 % of the variation obtained occur within populations. Although Mantel’s test had shown correlation between genetic distances and geographic was observed that Ivatuba and Itambé (33.8 km) have the small geographical distance and the largest genetic distance. The lower genetic distance was estimated for Maringá and Ivatuba but this localities have a small geographic distance
(42.3 km), indicating that there are no barriers for mating among reproducers in these populations. The high degree of polymorphism (83.11 %) and the ability to cross among the populations in the studied regions indicate that this species of leaf-cutting ant is well adapted to the region; therefore, integrated control programs can be developed.
Keywords: genetic variability, leaf-cutting, PCR-RAPD.
RESUMEN
La variabilidad genética de las hormigas Atta sexdens rubropilosa colectadas en cinco lugares distintos de Brasil fueron evaluados por la técnica PCR-RAPD. Un total de 15 primers produjeron 148 fragmentos, de los cuales 123 fueron polimórficos, lo que corresponden al 83,11 %. La estimación de la diversidad genética por el índice de Shannon fue 0,3836 y el desviación estándar fue de ± 0,2335. Estos valores demuestran una alta diversidad genética. El valor de GST fue 0,2372 y ΦPT = 0,184 lo que indica que las poblaciones están moderadamente diferenciadas y que el 82 % de la variación obtenida se produce dentro de las poblaciones. Aunque la prueba de Mantel ha demostrado una correlación entre la distancia genética y geográfica se observó que Ivatuba e Itambé (33,8 km) tiene una pequeña distancia geográfica y la mayor distancia genética. La distancia inferior genética fue estimada para Maringá e Ivatuba pero estas localidades cuentan con una distancia geográfica pequeña (42,3 km), lo que indica que no hay barreras para el apareamiento entre los reproductores en estas poblaciones. El alto valor de polimorfismo (83,11 %) y la capacidad de emparejamiento entre las poblaciones presentes en las regiones estudiadas, indican que esta especie de hormiga cortadora está bien adaptada a la región, y deben ser desarrollados programas integrados de control si se convierten en plagas.
Palabras clave: hormigas cortadoras, PCR-RAPD, variabilidad genética.
INTRODUCTION
Leaf-cutting ants are considered important pests in the neotropics because they have a wide geographical distribution and are responsible for cutting a wide variety of native and cultivated plants. In particular, Atta sexdens rubropilosa species (Forel, 1908) harm Eucalyptus and Pinus spp. (Boaretto et al., 1997) by cutting leaves and tender branches and are capable of completely destroying plants (Oliveira, 1996). This occurs mainly in homogeneous stands, which favor the development of pests, such as ants of the Atta and Acromyrmex genera (Della Lucia, 1993).
Forestry companies have used chemicals to control leaf-cutting ants. However, economic and environmental factors have led companies to improve the operational efficiency of chemical control techniques. In recent years, there has been an increase in the number of jobs related to biological and cultural control, especially in relation to plant resistance, in an attempt to identify alternatives to chemical control strategies or formulate combinations of different control strategies (Della Lucia, 1993).
Ants also have a variety of ecological functions as seed dispersers, contributing to the reforestation of many ecosystems and promoting their germination, removed the fruit pulp (Peternelli et al., 2004). They are responsible for pruning some plants, promoting their vegetative growth and play an important role in soil aeration (Hölldobler and Wilson, 1990), and incorporate organic matter on earth, making it fertile (Moutinho et al., 2003). In general, ants provide the unique opportunity to investigate various biological, evolutionary, and population issues because they form highly complex colonies due to their diverse social classes. Ants may be used to conduct ecological and population genetic structure studies, and they also provide rich material for investigating kin selection and altruism (Brian, 1983; Sudd and Franks, 1987; Hölldobler and Wilson, 1990). Ants are haplodiploid organisms, as the males emerge from parthenogenesis and females from fertilized eggs. The occurrence of two ploidy levels can be used as an important tool for genetic analysis (Diehl et al., 2002).
Atta colonies usually consists a queen (mongyne) and thousands or millions of sterile workers of different sizes and shapes (Hölldobler and Wilson, 2009). The adults, with rare exceptions, are females and are divided into at least two castes: fertile females or queens, whose primary function is to egg laying and sterile females, which perform all the other colony activities, such as water collection and food, feeding the young and queen, construction and defense of the nest (Wilson, 1976). Males constitute an additional caste and usually appear only once a year, in mating season, which occurs through the completion of the nuptial flight (Hölldobler and Wilson, 2009). The queen, after three to eight fertilized by males during the mating flight, falls to the ground and removes their wings to begin building the new nest and male die after mating (Forti, 1985). Some authors have argued that diploid organisms have greater genetic variability than haplodiploid organisms (Pamilo et al., 1978; Falcão and Contel, 1990). Graur (1985), concluded that the level of sociality is more important than haplodiploidy in determining genetic diversity in hymenoptera.
Since the 1960s, isoenzyme analysis has been the dominant method used for the evaluation of genetic variation in natural populations. Other techniques are currently available for the study of genetic diversity, such as random amplification of polymorphic DNA analysis (RAPD), but all techniques have advantages and disadvantages (Ferreira and Grattapaglia, 1998). RAPD is based on the identification of polymorphic DNA randomly amplified using a single primer with an arbitrary sequence, which is capable of amplifying DNA sequences contained between two annealing sites (Welsh and Mcclelland, 1990). Although this technique uses random sequences, many studies have shown that it can be used to effectively identify insect populations (Lopes-da-Silva et al., 2004) and for genetic variability studies. Using isozyme polymorphisms (Cantagalli et al., 2010) analyzed the population structures of Atta capiguara (honeybeebrown ants) in the Tapejara region (PR) and found low genetic variability for this species. Thus, the RAPD technique was used in this study to analyze the genetic variability and population structure of the A. s. rubropilosa leaf-cutter ant to increase the understanding of ant population genetics and improve the management of this species.
MATERIALS AND METHODS
BIOLOGICAL SAMPLE COLLECTION
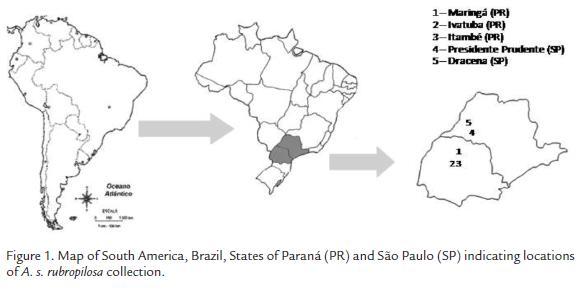
Atta sexdens rubropilosa soldiers, which are easily identified by their characteristic lemon grass smell due to a gland with an alarm function in the head, were collected from five different regions in the states of Paraná and São Paulo, Brazil. In northwestern Paraná, samples were collected from Maringá (23°26’ S, 51º56’ W), Itambé (23°39’39” S, 51°59’24” W), and Ivatuba (23°37’08” S, 52°13’15 W). In São Paulo, the samples were collected from Presidente Prudente (22°07’04’’ S and 51°22’57’’ W) and Dracena (21°28’57” S, 51°31’58” W). Collection localities can be visualized in Fig. 1. In Maringá, Itambé, and Ivatuba, four nests were sampled at each location, and in Presidente Prudente and Dracena, two nests were sampled per site, for a total of 16 nests. The collected material was stored in plastic bottles at -20 °C.
DNA ISOLATION
Total DNA was extracted from five individuals from each nest, with the exception of one nest from Ivatuba, where it was only possible to extract DNA from four individuals, for a total of 79 samples. Only the heads of soldiers were used to prevent contamination with nucleases that are present in the digestive tract of these insects. The method used for total DNA extraction was a modified version of that described by (Yu et al., 1993). The heads were homogenized in 1,5 ml microcentrifuge tubes containing 300 µl of extraction buffer [200 mM Tris-HCl (pH 8,0), 0,5 % SDS, 250 mM NaCl, 50 mM EDTA, and 100 mg/ml Proteinase K]. After 30 min of incubation at 65 °C, the material was centrifuged for 10 min (10,000 x g) and the supernatant was transferred to a new tube. An equal volume of chloroform: isoamyl alcohol (24:1) was then added. After homogenization and centrifugation for phase separation, the upper phase was transferred to a new tube. The DNA was precipitated with 250 µl of cold isopropanol and incubated at 20 °C overnight. The precipitated DNA was separated by centrifugation at 10,300 x g for 10 min. After discarding the supernatant, the precipitate was washed with 1.0 ml of cold 70 % ethanol and left to dry at room temperature. After drying, the DNA was resuspended for two hours in 50 µl of TE buffer, treated with RNAse (10 mg/ml), and incubated at room temperature for more than 2 hours before storage at -20 °C.
The DNA quality was evaluated and quantified on 0.8 % agarose gels with 1X TAE buffer. The amount of DNA present in each sample was estimated by comparison with known concentrations of a graded DNA standard (λ phage). The gels were stained in ethidium bromide bath (0.5 mg/mL), and the gel images were visualized under ultraviolet light and captured with the EDAS system (Kodak 1D Image Analysis 3.5, New York, USA).
AMPLIFICATION OF FRAGMENTS VIA PCRRAPD, SEPARATION, AND VISUALISATION OF PRODUCTS
To evaluate the genomes of the Atta sexdens rubropilosa populations, 15 primers were produced by Operon Technologies Inc., Alameda, CA, USA (OPA-01, OPA-02, OPA-03, OPA-04, OPA-07, OPA-10, OPA-11, OPA-14, OPA-18, OPA-19, OPB-11, OPB-12, OPB-17, OPC-20, and OPM-3). The amplification conditions were based on the methodology described by (Williams et al., 1990). For a reaction volume of 20 µl, it was used 1X Tris-KCl [20 mM Tris-HCl (pH 8.4) and 50 mM KCl], 2.5 mM MgCl2, 0.3 mM of primer, 0.1 mM each dNTP, one unit of Taq DNA polymerase (Invitrogen), and 20 ng of template DNA. Reactions were performed in an Eppendorf Mastercycler® Gradient thermocycler, as follow 96 °C for five minutes followed by 45 cycles of 30 seconds at 94 °C, 45 seconds at 35 °C, and one minute at 72 °C, with a final extension at 72 °C for seven minutes. A negative control reaction (with no DNA template) was used. The amplified products were separated in a 1.7 % agarose and stained in an ethidium bromide bath. The DNA fragments were visualized under an ultraviolet light, and the images were captured with the EDAS system (Kodak 1D Image Analysis 3.5).
POPULATION GENETICS ANALYSIS
The data analysis involved comparison of the molecular weights of the fragments (bands) obtained from the genomic DNA. Individual bands with identical molecular weights (located on the same loci) were compared within and between populations and designated as present (1) or absent (0) for each individual analyzed. Genetic variability within and between populations was determined by the percentage of polymorphic loci. The genetic diversity of the populations studied was estimated with the Shannon index (I) (Lewontin, 1972), GST values were also calculated. Nei’s (1978) genetic distance had been used to UPGMA cluster analysis (unweighted pair-group method using an arithmetic average) and construct a dendrogram. For these analyses, the Popgene 1.31 program was used (Yeh et al., 1999).
GenALEX 6.5 (Peakall and Smouse, 2012) software was used to estimate AMOVA (analysis of molecular variance) and phi’PT (ΦPT); to construct genetic relationships tree among A. s. rubropilosa populations based on Nei and Li/Dice; to estimate percentage confidence level in the bootstrap analysis (1,000 replicates); spatial analyses involving binary data were performed estimating principal coordinate analysis (PCA) and Mantel test (1967).
RESULTS
The amount of extracted DNA was ranged between 10 and 40 ng. To verify the genetic variability of Atta sexdens rubropilosa species collected from different region and the ability to amplify the extracted DNA via PCR-RAPD, 80 primers were initially tested, of which 15 were used. Table 1 shows the nucleotide sequences of the selected primers, the numbers of the total and polymorphic fragments, and the sizes of the amplified fragments.
The number of clear and reproducible fragments generated by the primers in all populations studied ranged from four to 17 fragments, with an average of 9.8 fragments per primer, and the sizes of the amplified products were between 200 and 5000 bp (Fig. 2). The 15 primers used in the RAPD-PCR reactions produced 148 fragments. It was observed that of out the total number of fragments analyzed, 123 (83.11 %) displayed polymorphisms.
The estimated genetic diversity obtained with the Shannon index (0.3836) and the standard deviation (± 0.2335) indicated that these populations had high genetic diversity. The high value of GST (0.2372) and ΦPT = 0.184 (significant probability at 0.010) showed that A. s. rubropilosa populations are moderately differentiated. GST can be considered an estimate of FST when it is assumed subpopulations are in Hardy-Weinberg equilibrium (FIS = 0). Therefore, considering Wright (1984) can be considered that GST values between 0.15 and 0.25 show that populations are moderately differentiated. AMOVA analysis showed that 82 % of the variation obtained with RAPD markers occurs within populations and 18 % among populations.
Principal Component Analysis (PCA) estimated by the covariance matrix showed that 71.84 % of the variation is explained by the coordinates 1 and 2 (Fig. 3). The first three coordinates explained 81.30 % of the variation. The coordinates 1, 2 and 3 correspond to 58.51 %, 13.33 % and 9.46 % respectively of variation.
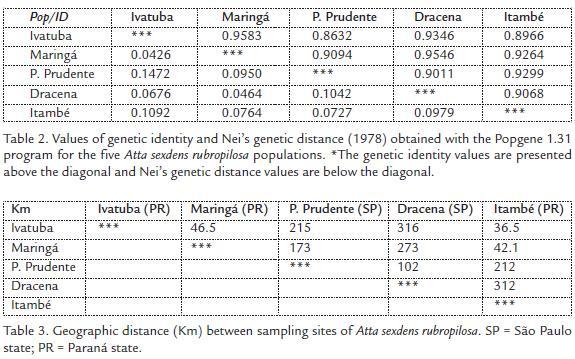
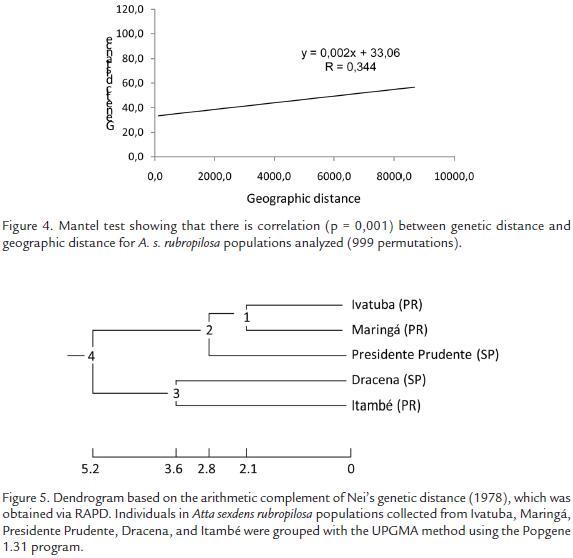
Nei’s genetic distance values among the five populations analyzed indicated that the populations from Presidente Prudente and Ivatuba had highest genetic distance (Table 2) but not geographical (215 km, Table 3), whereas the populations from Maringá and Ivatuba had the lowest genetic distance (Table 2) but not geographical (46.5, Table 3). The correlation between genetic distance and geographical distance was evaluated by Mantel’s test. Fig. 4 shows that there is correlation between these variables (R = 0.344, p = 0.001). A dendrogram based on the arithmetic complement of Nei’s genetic distance (1978) (Fig. 5) shows that A. s. rubropilosa analyzed can be separated into two main groups. The first is formed by Ivatuba-PR, Maringá-PR and Presidente Prudente-SP, the second has populations from Dracena-SP and Itambé-PR (Fig. 5).
UPGMA clustering based on the arithmetic complement of the Jaccard coefficient was not used because the values were not significant at the 70 % level by bootstrap analysis with 1,000 repetitions.
DISCUSSION
Carvalho and Vieira (2001) reported that the amount of DNA in relation to the primer is one of the key factors for successful PCR because amplification may not occur if the DNA quantity is too low or high. Carvalho (2000) found that for A. s. rubropilosa, the optimum amount of DNA for RAPD reactions is between 19 and 30 ng. Our study shows that it was possible to extract highquality DNA that ranged between 10 and 40 ng for amplification.
RAPD-PCR marker proved efficient for genetic populations studies of A. s. rubropilosa, because 148 fragments reproducible were obtained with 15 primers. Grutzmacheri et al. (2007) used three primers (UBC 354, UBC 348, and UBC356) for RAPD to study the genetic variability of the Acromyrmex genus collected from Rio Grande do Sul and obtained 87 fragments. Leaf-cutting ants of the Atta genus are indigenous to the Americas. In the Americas, the distribution of these insects extends from the southern U.S. to northern Argentina, except for Chile and some of the Antilles islands (Sousa, 1996). Originally, there were no Atta ants in the forests of Paraná. They colonized the state following the first reforestation event in Paraná during the 1940s (Bansho et al., 1994).
In the short time since the introduction of A. s. rubropilosa ants in Paraná (approximately 70 years), they have obtained a high degree of differentiation (GST = 0.2372), but there is high variation within populations as shown by AMOVA. Chemical control of A. s. rubropilosa populations considered pests by farmers, should be careful. The high intrapopulation polymorphism and possible gene flow between populations may promote the occurrence of cross-resistance to pesticides used to control them.
Despite the short geographical distance (36.5 km) between Ivatuba and Itambé, these populations have the highest genetic distance (Table 2). It is possible to infer that these breeding populations do not mate or that there are a small number of hybridizations. Among the Maringá and Ivatuba populations (separated by 42.3 km), the lowest estimated genetic distance was observed (Table 3). However Mantel test (R = 0.344; p = 0.001) indicate that there is relationship between the genetic and geographic distances of the studied nests. These results support stepping-stone model proposed by Kimura (1953). This model assumes that species occupying a wide geographic area, forming differentiate geographical races (Kimura and Weiss, 1964). Analyzed A. s. rubropilosa populations do not constitute races, but can be considered subpopulations. The high genetic differentiation values may be due to the existence of an abundant ancestral polymorphism that can be maintained by multiple mating of the queen. The high polymorphism (83.11 %) indicates that this species of leaf-cutter ant is adapted to the region and integrated programs for the control should be developed.
CONCLUSIONS
Leafcutting ants’ A. s. rubropilosa showed extensive polymorphism and genetic diversity that were higher within populations than between them. The values of genetic distance and geographic are not randomized and analyzed populations are moderately differentiated.
ACKNOWLEDGEMENTS
Thanks to Post Graduation in Genetic and Breeding (State University of Maringá/Paraná State/Brazil) and to CAPES Program (Coordination Improvement Higher Education Personnel).
BIBLIOGRAPHY
BANSHO JY, CARNEIRO DA, CORDEIRO L. Controle de formigas cortadeiras na KFPC – PR. Anais do III Curso de Atualização no Controle de Formigas Cortadeiras – IPEF; 1994. p. 41-50.
BOARETTO MA, FORTI LC. Prospects in control of leaf-cutting ants. Série Técnica, IPEF. 1997;11:31-46.
BRIAN MV. Social Insects: ecology and behavioral biology. New York: Chapman and Hall; 1983. p. 377.
CANTAGALLI LB, MANGOLIN CA, RUVOLOTAKASUSUKI MCC. Isoenzymatic Polymorphism in the Leaf-Cutting Ant Atta capiguara Gonçalves (Hymenoptera: Formicidae). Neotrop Entomol. 2010;39(1):46-49.
pCARVALHO AOR, VIEIRA LGE. Determination of optimum conditions for PCRRAPD analyses in Atta sexdens rubropilosa Forel (Hymenoptera:Formicidae). Londrina: Neotrop Entomol. 2001;30(4):593-600.CARVALHO AOR. Analysis of genetic variability and identification of species of the Atta (Hymenoptera: Formicidae) genera via molecular markers. [Doctoral Thesis]. Curitiba: Federal University of Paraná; 2000. p. 134.
DELLA LUCIA TMC. Leaf-cutter ants. Folha de Viçosa; 1993. p. 262.
DIEHL E, CAVALIMOLINA S, ARAÚJO AM. Isoenzyme variation in the leaf-cutting ants Acromyrmex hayeri and Acromyrmex striatus (Hymenoptera, formicidae). Genet Mol Biol. 2002;25(2):173-178.
FALCÃO TMMA, CONTEL EPB. Genetic variability in natural populations of Brazilian social bees. I. Isozyme patterns and polymorphism for esterases and total protein. Braz J Genet. 1990;13(4):731-754.
FERREIRA ME, GRATTAPAGLIA DIntroduction to the use of molecular markers in genetic analysis. Brasília: Embrapa-Cenargen; 1998. p. 220.
FORTI LC. Ecologia da saúva, Atta capiguara Gonçalves, 1944 (Hymenoptera: Formicidae) em pastagens. Tese (Doutorado em Ciências). Piracicaba: Escola Superior de Agricultura Luiz de Queiroz; 1985. p. 234.
GRAUR D. Gene diversity in Hymenoptera. Evolution. 1985;39(1):190-199.
GRUTZMACHERI DD, LOECK AE, OLIVEIRA AC, ZIMMER PD, MALONE G. Interspecific genetic diversity in leaf-cutter ants of the Acromyrmex genera that occur in the state of Rio Grande do Sul. Ciência Rural. 2007;37(4):921-927.
HÖLLDOBLER B, WILSON EO, The super-organism. Cambridge, Mass: Harvard University Press; 1990. p. 732.
HÖLLDOBLER B, WILSON EO, The super-organism. Morton & Company; 2009. p. 522.
KIMURA M. Population genetics, molecular evolution, and the neutral theory: selected papers. University of Chicago Press; 1994. p. 133.
KIMURA M, WEISS GH. The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49(9):561-576.
LEWONTIN RC. The apportionment of human diversity. Evol Biol. 1972;6:381-398.
LOPES-D-SILVA M, TONET GEL, VIEIRA LGE. Characterization and genetic relationships among Brazilian biotypes of Schizaphis graminum (Rondani) (Hemiptera: Aphididae) using RAPD markers. Londrina. Neotrop Entomol. 2004;33(1):43-49.
MANTEL N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27(2):209-220.
MOUTINHO P, NEPSTAD DC, DAVIDSON EA. “Influence of leaf-cutting ant nests on secondary Forest growth and soil properties in Amazônia”. Washington. Ecology. 2003;84(5):1265-1276.
NEI M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89(3):583-590.
OLIVEIRA MA. Identificação de formigas cortadeiras e efeito do desfolhamento simulado em plantios de Eucalyptus grandis. Dissertação (Mestrado em Entomologia), Universidade Federal de Viçosa, Viçosa; 1996. p. 67.
PAMILO P, ROSENGREN R, VEPSÄLÄINEN K, VARVIO-AHO SL, PISARSKI B. Population genetics of Formica ants. I. Patterns of enzymes gene variation. Hereditas. 1978;89(2):233-248.
PEAKALL R, SMOUSE PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics. 2012; 28(19):2537-2539.
PETERNELLI EFO, DELLA LUCIA TMC, MARTINS SV. Espécies de formigas que interagem com as sementes de Mabea fistulifera Mart. (Euphorbiaceae). R Árvore. 2004;28(5):733-738.
SOUSA NJ. Evaluation of use of three types of doorbaits to control ants in areas prepared for settlement deployment of Pinus taeda L. [Magister Thesis].Curitiba: Federal University of Paraná, 1996.
SUDD JH, FRANKS NR. The behavioral ecology of ants. New York: Chapman and Hall; 1987. p. 206.
WELSH J, MCCLELLAND M. Fingerprinting genomes using PCR with arbitrary primers. Nucl Acids Res. 1990;18(24):7213-7218.
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18(22):6531-6535.
WILSON EO. A social ethogram of the neotropical arboreal ant Zacryptocerus varians (Ft. Smith). Anita Behav. 1976;24:354-363.
WRIGHT S. Evolution and the genetics of populations, volume 4, variability within and among natural populations. The University of Chicago Press, LTD.; 1984. p. 580.
YEH FC, BOYLE TYZ, XIYAN JM. POPGENE Version 1.31: Microsoft Windowbased freeware for population genetic analysis. Edmonton: University of Alberta and Center for International Forestry Research; 1999.
YU KF, VAN DEYNZE A, PAULUS KP. Random amplified polymorphic DNA (RAPD) analysis. In: BR Glick (ed.). Methods in plant molecular biology and biotechnology. USA: CRC Press; 1993. p. 287-301.
Cómo citar
APA
ACM
ACS
ABNT
Chicago
Harvard
IEEE
MLA
Turabian
Vancouver
Descargar cita
Visitas a la página del resumen del artículo
Descargas
Licencia
Derechos de autor 2013 Acta Biológica Colombiana

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
1. La aceptación de manuscritos por parte de la revista implicará, además de su edición electrónica de acceso abierto bajo licencia Attribution-NonCommercial-ShareAlike 4.0 (CC BY NC SA), la inclusión y difusión del texto completo a través del repositorio institucional de la Universidad Nacional de Colombia y en todas aquellas bases de datos especializadas que el editor considere adecuadas para su indización con miras a incrementar la visibilidad de la revista.
2. Acta Biológica Colombiana permite a los autores archivar, descargar y compartir, la versión final publicada, así como las versiones pre-print y post-print incluyendo un encabezado con la referencia bibliográfica del articulo publicado.
3. Los autores/as podrán adoptar otros acuerdos de licencia no exclusiva de distribución de la versión de la obra publicada (p. ej.: depositarla en un archivo telemático institucional o publicarla en un volumen monográfico) siempre que se indique la publicación inicial en esta revista.
4. Se permite y recomienda a los autores/as difundir su obra a través de Internet (p. ej.: en archivos institucionales, en su página web o en redes sociales cientificas como Academia, Researchgate; Mendelay) lo cual puede producir intercambios interesantes y aumentar las citas de la obra publicada. (Véase El efecto del acceso abierto).