IDENTIFICATION OF KEY MOLECULAR COMPONENTS OF THE RESISTANCE OF CHERRY TOMATO AGAINST Phytophthora infestans
Palabras clave:
Compatible interaction, incompatible interaction, Matt´s wild cherry, Solanum lycopersicum var. cerasiforme, Phytophthora infestans. (es)
Cherry tomato Solanum lycopersicum var cerasiforme cv Matt’s wild cherry is a very resistant cultivar to most Phytophthora infestans isolates. Two isolates were identified, US940480 and US970001 that cause an incompatible and a compatible interaction respectively. US970001 is one of the few isolates producing a compatible interaction with this cultivar. To identify genes with a differential gene expression between compatible and incompatible interactions, gene expression patterns were analyzed with tomato cDNA microarrays including 12,899 independent tomato cDNA clones at different time points after inoculation. A diverse set of statistical tools were used to identify key components of the plant response to the pathogen. Forty-three genes were up-regulated during the incompatible reaction at time point 36 hours, 15 globally at all time points and twelve were found both in globally and at 36 hours. Northern blots analysis was performed to confirm differential expression showed by microarray analysis and to study the differential expression of more PR genes between compatible and incompatible interactions for this interaction.
IDENTIFICATION OF KEY MOLECULAR COMPONENTS OF THE RESISTANCE OF CHERRY TOMATO AGAINSTPhytophthora infestans
Identificación de los principales componentes moleculares de la resistencia de tomate cherry contra Phytophthora infestans
LILIANA LÓPEZ KLEINE1, Ph. D.; CHRISTINE D. SMART2, Ph. D.; WILLIAM E. FRY2, Ph. D.; SILVIA RESTREPO3, Ph. D.
1Departamento de Estadística, Universidad Nacional de Colombia, Bogotá, Colombia. llopezk@unal.edu.co 2Department of Plant Pathology and Plant-Microbe Biology, Cornell University, USA. Fry: wef1@cornell.edu; Smart:dcs14@cornell.edu. 3Departamento de Ciencias Biológicas, Universidad de los Andes, Bogotá, Colombia. Corresponding: srestrep@uniandes.edu.co
Presentado el 30 de julio de 2012, aceptado el 23 de octubre de 2012, correcciones el 1 de noviembre de 2012.
ABSTRACT
Cherry tomato Solanum lycopersicum var cerasiforme cv Matt's wild cherry is a very resistant cultivar to most Phytophthora infestans isolates. Two isolates were identified, US940480 and US970001 that cause an incompatible and a compatible interaction respectively. US970001 is one of the few isolates producing a compatible interaction with this cultivar. To identify genes with a differential gene expression between compatible and incompatible interactions, gene expression patterns were analyzed with tomato cDNA microarrays including 12,899 independent tomato cDNA clones at different time points after inoculation. A diverse set of statistical tools were used to identify key components of the plant response to the pathogen. Forty-three genes were up-regulated during the incompatible reaction at time point 36 hours, 15 globally at all time points and twelve were found both in globally and at 36 hours. Northern blots analysis was performed to confirm differential expression showed by microarray analysis and to study the differential expression of more plant resistance genes (PR) genes between compatible and incompatible interactions for this interaction.
Keywords: Compatible interaction, incompatible interaction, Matt's wild cherry, Solanum lycopersicum var. cerasiforme, Phytophthora infestans.
RESUMEN
El tomate cherry Solanum lycopersicum var cerasiforme cv Matt's es bastante resistente a la gran parte de aislamientos de Phytophthora infestans. Se han identificado dos aislamientos, US940480 y US970001 que causan interacción incompatible y compatible respectivamente. US970001 es uno de los pocos aislamientos causantes de interacción compatible con este cultivo. Con el fin de identificar genes con expresión diferencial en interacciones compatible e incompatible, analizamos DNA copia de 12899 clones independientes en tres tiempos posteriores a la inoculación del patógeno. Se aplicaron diversas herramientas estadísticas para identificar componentes moleculares claves de la respuesta de la planta al patógeno. Cuarenta y tres genes fueron detectados como activados durante la interacción incompatible a las 36 horas posinoculación, 15 genes se detectaron como activados globalmente tomando en conjunto los 3 tiempos analizados y 12 genes tanto globalmente como a las 36 horas. Análisis de Northern blot permitieron confirmar la expresión diferencial detectada con los análisis de microarreglos y estudiar la expresión diferencial de otros genes de resistencia en plantas (PR) en in teracciones compatible e incompatible en esta interacción.
Palabras clave: interacción compatible, interacción incompatible, tomate cherry Matt's, Solanum lycopersicum var. cerasiforme, Phytophthora infestans.
INTRODUCCIÓN
Three well-defined types of host-pathogen interactions between tomato and Phytophthora infestans occur: highly compatible, partially compatible and incompatible interactions (Gallegly and Marvel, 1995). Several studies have focused in the partial compatible and compatible interaction between tomato and Phytophthora infestans. A previous study showed that partial compatibility in tomato against P. infestans acts independently of ethylene, Salicylic acid (SA), and Jasmonic Acid (JA) defense response pathways (Smart et al., 2003). Other studies showed that compatibility is dependent upon salicylic acid (SA) and ethylene but apparently is not dependent upon the jasmonic acid (JA) signaling pathways (Niderman et al., 1995; Fidantsef et al., 1999; Stout et al., 1999). Additionally, it is possible that patterns of ethylene-responsive plant resistance gene (PR) expression may be a general response to the biotrophic nature of highly compatible interactions between P. infestans and tomato rather than a specific defense response (Smart et al., 2003). However, recent studies that could shed light to this matter and the involvement of other metabolic pathways in the compatibility between P. infestans and tomato are not found.
Highly compatible and partially compatible interactions were studied using the susceptible tomato cultivar Rio Grande inoculated with either a tomato-specialized isolate (highly compatible) or a non-specialized isolate (partially compatible) (Smart et al., 2003). As expected, there was induction of the hypersensitive response (HR) earlier during the partially compatible interaction. However, contrary to our expectation, pathogenesis-related (PR) gene expression was not stimulated sooner in the partially compatible interaction.
In a general sense, the interaction between tomato and its pathogens is complex and besides, the aforementioned signaling pathways, it involves the co-regulation of gene expression, photosynthesis and sugar levels (Berger et al., 2004). The repression of photosynthesis has been observed in cell suspension cultures of tomato treated with elicitors (Sinha et al., 2002), in intact plants after the interaction with viral (Herbers et al., 2000; Hanssen et al., 2011), bacterial (Kocal et al., 2008) and fungal pathogens (Prokopová et al., 2010) although in the latter case the impairment of photosynthesis was minimal. The more common explanation to this change in metabolism is the switch from normal to defense metabolism when a plant is challenged by a pathogen. In potato, down-regulation as a consequence of the P. infestans compatible interaction was observed for genes encoding proteins involved in photosynthesis (Restrepo et al., 2005). The reason for the reduction in photosynthesis during the compatible interaction between potato and P. infestans is currently unknown.
Now, compatible interaction between an isolate of P. infestans (US970001) and a cherry tomato S. lycopersicum var cerasiforme cv Matt's wild cherry (Muller, 1940) were identified. This is a very resistant cultivar, which is only partially compatible with even the most aggressive tomato-specialized isolates (such as US980025) and incompatible with the isolate US940480. These interactions on Matt's wild cherry do allow us to compare host resistance responses between compatible and incompatible interactions.
MATERIALS AND METHODS
PLANTS, PATHOGEN ISOLATES, AND INOCULATIONS
Four-week-old Solanum lycopersicum var cerasiforme cv Matt's wild cherry plants were placed into a humid chamber with a 16 h light period, 12 h per day of 100 % relative humidity, and a temperature of 15 °C. Plants were inoculated with P. infestans isolates US940480 (ATCC # 208834, a member of the US-8 clonal lineage) resulting in an incompatible interaction, or with isolate US970001 (ATCC # MYA-2350, a member of the US-17 clonal lineage) resulting in a compatible interaction. Another group of plants was sprayed with water (the mock-inoculated control). Inoculations were made with a sporangial suspension of 20,000/mL, and plants were sprayed until run-off as previously described (Smart et al., 1998). Tissue (all leaflets) from three plants per group was collected at three time points (12, 36 and 72 hours after inoculation). The leaflets for each treatment at each time-point were pooled, flash frozen in liquid nitrogen, and stored at -80 °C. After the 72 h time-point, there were two plants left from each group (incompatible, compatible and mock-inoculated). These plants were kept in the humid chamber for an additional seven days to ensure that plants inoculated with the compatible isolate (US970001) became fully diseased, while those inoculated with the incompatible isolate (US940480) or mock-inoculated with water remained healthy. The entire experiment was repeated three times.
Tomato RNA isolation and preparation of array probes. Frozen plant tissues were ground using a cold mortar and a pestle. RNA was isolated from healthy tomato tissue, inoculated tomato tissue or in vitro grown P. infestans, using a previously described hot phenol method (Perry and Francki, 1992) with modifications described by Gu et al., (2000). Ten µg of total RNA from each sample was separated electrophoretically on a 1.2 % formaldehyde-agarose gel, and transferred to Hybond-N membrane (Amersham Biosciences, Piscataway, NJ). Hybridizations were performed using Puregene hyb-9 hybridization solution (Gentra Systems, Plymouth, MN). The RNA was precipitated, pooled and stored at -80 °C.
For each array hybridization, cDNA was generated from total RNA (150 µg) isolated from Matt's wild cherry tomato leaflets inoculated with P. infestans, isolates US940480 (incompatible interaction) or US970001 (compatible interaction). cDNA from the incompatible interaction was labeled with one cyanine fluorophore (Cy3 or Cy5), while cDNA from the compatible interaction was labeled with the other. All protocols used to generate and label cDNA were as described by Hedge et al (Hedge et al., 2000).
MICROARRAY HYBRIDIZATION AND SCANNING
Tomato cDNA arrays generated by the Center for Gene Expression Profiling (CGEP), Boyce Thompson Institute, Ithaca, New York, USA were used in these experiments. All clones, more than 12,800, were validated through resequencing and agarose gel electrophoresis prior to printing to confirm the sequence of the clone and the presence of an insert. All data on the EST sequences, the clones on the array, and annotation of the clones can be found at the Tomato Functional Genomics Database (TFGD) web site at http://ted.bti.cornell.edu/ Arrays were probed with infected tomato tissue (incompatible and compatible interaction) cDNA at each of the three time-points (12, 36 and 72 hpi). Two arrays per time point (3) were hybridized using the dye-swap design for each of the three biological replicates for a total of 18 arrays. Additionally, one dye swap experiment before infection was undertaken (2 arrays).
Arrays were pre-hybridized in order to block nonspecific background during hybridization. Slides were blocked in 5 X SSC, 0.1 % SDS, and 1 % bovine serum albumin at 42 °C for 45 min (Hedge et al., 2000). Slides were washed in sterile distilled water followed by isopropanol and dried. Cy-3 and Cy-5 probes were combined, placed on the slide, and covered using a glass cover slip washed in 1 % SDS. Arrays were put into hybridization chambers (Corning Inc, NY) and hybridized overnight at 42 °C in a water bath. The slides were removed from the chambers and washed in 2 X SSC and 0.1 % SDS at 42 °C from 5 min., in 0.1 X SSC and 0.1 % SDS at room temperature for 5 min., and twice in 0.1 X SSC at room temperature for 5 min., then dried. Slides were scanned using an Axon GenePix 4100 microarray scanner (Axon Instruments Inc, Union City CA). The photomultiplier settings were adjusted in each channel to result in non- saturation of the most highly expressed genes.
ARRAY DATA ANALYSIS
To determine fluorescence intensity and background intensity, 16-bit TIFF scanned images were analyzed using the software GenePix Pro version 4.1 (Axon Instruments Inc). The median pixel intensity within a given spot was used as the signal intensity. For the Cy5 dye, the background signal intensity determined by GenePix was occasionally higher than that of genes that were not highly expressed but that had previously been shown to be differentially expressed (Smart et al., 2003). Therefore, to avoid negative values we utilized the strategy proposed by Frick and Schaller, (2002), and did not subtract the background signal. The log2-transformed expression values across all samples were standardized to a mean of zero and standard deviation of one (row standardization). Differentially expressed genes were determined using significance analysis of microarray (SAM) proposed by Tusher et al., (2001). At each time-point, differentially expressed genes were detected comparing mean expression in each of the two conditions (incompatible and compatible interactions). An overall comparison of both conditions was done comparing mean expression of each condition along all time-points. This global comparison could mask genes that are up-regulated at one time- point and down-regulated at another time point but allows the detection of genes that are consistently either up-or down-regulated.
DATABASE SUBMISSION OF MICROARRAY DATA
The microarray data were prepared according to the Minimum Information about a Microarray Experiment (MIAME) recommendations (Brazma et al., 2001) and submitted to the TED (Tomato Expression Database) database which is available at http://ted.bti.cornell.edu/.
PRINCIPAL COMPONENT ANALYSIS
A Principal Component Analysis (PCA) was conducted using genes as individuals and all time point hybridizations as variables by means of the ade4 R package (Dray et al., 2007). The result of the PCA is the definition of new variables called Principal Components (PCs) that are linear combinations of the original variables and allow representing graphically in less dimensions (usually in two) the overall phenomena and data structure. Here the first two PC were used to represent the original 40 microarray conditions. The 40 microarray conditions correspond to 40 total RNAs hybridized: three time points, three biological replicates, two dye swaps per time point and two self-self hybridizations, a total of 20 hybridations, 40 RNAs, two RNAs (a Cy3 and a Cy5 - labeled RNA) are combined per hybridization.
PCA also produces the coordinates of all individuals (here genes) in the new two- dimensional space (plane). These coordinates are locations of the genes on a plane and can be used to plot genes on this space. Coordinates on the PC space of genes were used to characterize similarities between gene sets involved in resistance and disease processes. Based on previous studies we constructed a set of 187 genes involved in resistance and disease processes, see next section. The projection of genes on the PC space was plotted highlighting each of the gene sets with a different color and indicating their distance to the centroid (mean point) of each group. Moreover, the distances between genes of each group were obtained on the PC space of the first two components in order to compare the distance between and within groups.
CONSTRUCTION OF GENE SETS IMPLICATED IN RESISTANCE PROCESSES
Based on previous studies we constructed a set of 187 genes involved in resistance and disease processes. A total of 143 genes out of the 187 complete set, were filtered based solely in the gene description of the best BLAST hit and categorized as "Phytophthora inhibited protein", "Avr elicited" or "disease resistance". The remaining forty-four genes were classified as "inhibited by EPI1 and EPI2" (EPI: extracellular serine protease) (Tian et al., 2004; Tian et al., 2005; Tian et al., 2007), WRKYs (Eulgem T. 2006) and "Pathogenicity associated" (Restrepo et al., 2005).
NORTHERN BLOT ANALYSIS
DNA probes were labeled using the Random Primers DNA Labeling System according to the manufacturers' protocol (Invitrogen, Carlsbad, CA). The PR genes used as probes were identical to those described by Gu et al., (2000) and included; acidic glucanase (GluA), basic glucanase (GluB), basic PR-1, and divinyl ether synthase (DES) and carbonic anhydrase (CA).
RESULTS AND DISCUSSION
A LOW NUMBER OF EXPRESSED GENES DIFFERENTIATES A COMPATIBLE AND AN INCOMPATIBLE REACTION
Expression analysis revealed a very low number of differentially expressed genes between compatible and incompatible interactions on Matt's wild cherry to P. infestans. Analysis of gene expression on the tomato (cv Matt's wild cherry) - P. infestans interaction compared two interactions, compatible and incompatible. In this study ratios correspond to expression in an incompatible interaction divided by expression in a compatible interaction. No differentially expressed genes were detected at 12 h or 60 h after inoculation. A total of 43/12899 up-regulated genes in the incompatible reaction were detected at time point 36. Combining all time points together, 15 of the 12899 clones represented on the TOM1 microarray were found to be up-regulated in the incompatible interaction vs. compatible interaction and 12 genes were up-regulated both at 36 h and globally (combining all time points). The pattern of expression was clearer when we considered genes up-regulated globally and at 36 h in the incompatible interaction since at least four genes out of 12 were involved in oxygen and free-radical metabolism, and a PR coding for proteins known for their involvement in signal transduction were also detected (supplementary table 1). Among the up-regulated genes at 36h in the incompatible cultivar (down-regulated for the compatible interaction), we could find PR proteins, photosystem I associated proteins and different types of kinases (supplementary table 1).
Novel statistical analyses reveal key molecular components of defense. Variability of gene expression fold change ratio after log normalization was analyzed and it was possible to detect that the overall variability is very low (standard deviations are 0.22 for the global experiment, 0.56 for the -12 timepoint and 0.33, 0.74, 0.27 for the 12, 36 and 60 timepoints respectively). This can be seen graphically in the boxplots depicted in figure 1. Interestingly, the most variable treatment was the experiment at timepoint -12. The most possible explanation for this can be that only one experiment (only one biological replicate) was available and the fold change is not applicable because it is not a real comparison of two conditions. This hybridization was only performed as a self- self control, the hybridization of two identical RNAs samples. This indicated that the variability of gene expression in this whole experiment is low, suggesting very few changes in gene expression in this particular pathosystem between a compatible and an incompatible interaction.
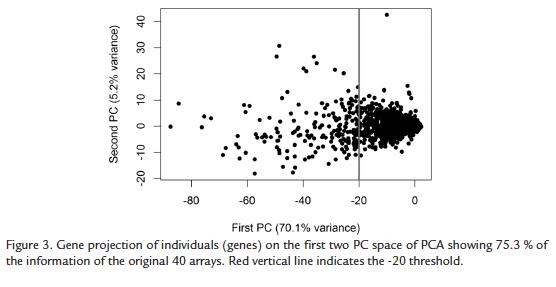
The PCA conducted on microarray data using genes as individuals was able to reduce the 40 original variables to two principal components (PCs) very successfully, as 75.3 % of the variance was retained in these first components. Moreover, the first PC retained 70.1 % of the variance indicating that most hybridizations are highly correlated and that differences in gene expression between time points is low or affects only few genes, which is on accordance with the low percentage of differentially expressed genes detected by SAM. As can be observed in figure 2 most variables are highly correlated with and therefore represented by PC1. It is worth mentioning that variables with most different behavior are V36 and V35, which correspond to replicates of time point 60. As differential expression was only detected at time point 36 between compatible and incompatible interactions, variability of gene expression at time point 60 is apparently not due to differential expression between these two conditions but to random variability due to differences between individuals at this time point.
Mean correlations of each one of the hybridizations with all other hybridizations are shown in table 1. As expected, correlations of V35 and V36 to the other hybridizations are lower (Table 1) as these variables representing expression at 60h showed a distinct behavior (Fig. 2). Moreover, correlations between all other experiments are around 0.7 confirming that all experiments are related even though they belong to different conditions (incompatible and compatible interaction) and time points, confirming again that very few genes are differentially expressed between a compatible and an incompatible interaction in this pathosystem.
When genes are plotted on the plane of the first two PCs, (which represent 75.3 % of the information contained in the whole data set), no apparent structure in the data was detected. The behavior of most genes is neither differentiated nor characteristic, showing expression values around zero (the mean expression value in normalized and centered data). Therefore most of the genes are placed around the origin of the PC space. Only few genes behave in a different way, mainly in relationship to the first PC, which means that they behave differently than most of the other genes. Nevertheless, these genes do not group in a separate cluster but each one has a unique behavior (Fig. 3). Among these genes we could detect some defense-, wound-, harpin and Avr9- induced genes and enzymes belonging to the ethylene synthesis pathway. However, of high interest was the finding in this set of differentially expressed genes has a high number of genes related to photosynthesis and to the Halliwell-Asada enzyme pathway: ascorbate oxidase, monodehydroascorbate reductase, glutathione peroxidase, superoxide dismutase, glutathione- S-transferase, Rubisco, chlorophyll a/b binding proteins and carbonic anhydrase among others. All these genes were up-regulated in the incompatible interaction or down-regulated in the compatible interaction confirming previous results in other P. infestans hosts (Restrepo et al., 2005).
For the purposes of this study, a set of genes involved in resistance and disease processes was built. Genes of interest and belonging to disease-associated gene-sets are highlighted in figure 4. Globally, all these genes behave similarly. However, two genes, one with WRKY motif (SGN-U214610; AAAR98818) and one inhibited by EPI1 or EPI10 (SGN-U212610; AAA80496) showed a strikingly different behavior.
VALIDATION OF DIFFERENTIAL GENE EXPRESSION BY NORTHERN BLOTS
Northern blots are the most reliable and robust analysis to confirm differential gene expression. Northern analysis confirmed the similarity of gene expression between a compatible and incompatible interaction in the Matt's wild cherry tomato. Expression patterns of PR genes and carbonic anhydrase (CA) in the compatible and incompatible interactions in tomato cultivar Matt's wild cherry were investigated. In general no differential expression was observed for the selected genes, confirming the microarrays results. For GluA (Acidic glucanase), a higher level of expression was observed during the compatible interaction at 60 h after inoculation. For GluB (Basic glucanase) and PR1b we could detect induction at 12 h after inoculation and more expression at 36 h for the compatible interaction. For DES (divinyl ether synthase), a transient induction at 12 h in both interactions was detected but at higher levels for the compatible interaction. Finally, for CA we observed an expression at high levels in healthy plants but repression after 36 h in the compatible interaction (Fig. 5).
COMBINING STATISTICAL AND BIOLOGICAL EVIDENCE TOWARDS THE ELUCIDATION OF DEFENSE MECHANISMS ON MATT'S CHERRY TOMATOES
From previous microarrays studies different groups have performed, it is now evident that more robust and creative ways of data analysis are needed to detect key genes in the evaluated treatments. It is known that the traditional statistical analyses of microarrays could discard interesting genes that do not show dramatically expression changes. Additionally, the appropriate combination of statistical analyses can help in the selection of genes to be functionally evaluated by different approaches including VIGS. This technology is not always successful in tomato, and even when it is successful it occurs as a mosaic (Rotenberg et al., 2006), so the careful selection of genes to be silenced will reduce general costs of the whole experiment. The success in finding the differences among the reactions (compatibility and incompatibility) and identifying the key molecular components of resistance was the use of a novel combination of several statistical tools. The most intriguing result obtained in this study was the almost lack of molecular differences between compatible and resistant reactions of the Matt's wild cherry tomato against Phytophthora infestans. Using a common microarray analysis, the Significance Analysis of Microarrays (SAM), we observed that differential gene expression was very late when comparing resistant and susceptible reactions of the Matt's wild cherry tomato against two genotypes of the pathogen P. infestans. No differentially expressed genes were detected before 36 hours after inoculations. Actually, no differences in gene expression between the two types of interactions were observed at 60 hours suggesting that molecular processes involved in resistance in the Matt's cherry tomato cultivar could be due to i) the genes detected at 36 h in this study (Supplementary table 1) or ii) to the activity of one major gene up-regulated very early during the interaction, before 12 hpi and complemented with the genes detected in this study or iii) to the activity of a gene or the activities of a set of genes not detected at the thresholds defined in our SAM microarray analysis.
We therefore used a novel combination of old statistical techniques trying to detect key components of defense not detected by SAM. The PCA indicated that most of the time points (here the variables) have correlated behaviors. This shows that differences between replicates and also between compatible and incompatible interaction conditions are globally very weak and that the differences in gene expression that exist are not enough for a change in data structure. Moreover, the projection of the genes on the PC space allows stating that most genes have very low expression and do not change this behavior along the conditions analyzed. This confirms the result of a low number of differentially expressed genes. However, PCA allowed the detection of a set of differentially expressed genes that had been identified in previous studies on potato late blight (Restrepo et al., 2005). As previously shown, the largest group of genes with a different behavior corresponds to photosynthesis-related genes (Schenk et al., 2000; Mysore et al., 2002; Gibly et al., 2004; Restrepo et al., 2005). These genes can be considered as down-regulated in the compatible interaction. As for the study of compatibility in potato challenged by P. infestans (Restrepo et al., 2005), results shown herein suggest that under stressful conditions, the alteration of expression of carbonic anhydrase impacts photosynthesis through the Halliwell-Asada pathway and glutamate metabolism (Restrepo et al., 2005; Pinzón et al., 2010). Then, during compatibility, other genes such as those encoding JA- pathway proteins in potato could be down-regulated and result in disease (Restrepo et al., 2005) but in resistant genotypes this effect might be rapidly counteracted. Regarding the known disease associated genes that we grouped in five gene sets, the PCA revealed that they do not have a particular behavior as a group but most of them are expressed weakly and do not change in the analyzed conditions. Only two genes showed a particular behavior, one with WRKY motifs and one inhibited by EPI1 or EPI10. Again, the involvement of these genes in defense or compatibility has to be functionally validated even if the WRKY role in resistance is very well documented as a positive and negative regulator.
CONCLUSIONS
In conclusion, very few differences between the gene induction in the susceptible and resistant interactions between Matt's wild cherry tomato and its pathogen P. infestans could be observed. Lack of marked differences in gene expression was also observed for the signaling pathways. In previous studies, it was shown that differences between compatible and incompatible interactions can be mainly explained quantitatively (Tao et al., 2003). However, a novel combination of statistical tools helped us to identify the key components of resistance in a tomato genotype showing interesting levels of resistance to its pathogens.
REFERENCES
BERGER S, PAPADOPOULOS M, SCHREIBER U, KAISER W, ROITSCH T. Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol plantarum. 2004;122(4):419-428.
BRAZMA A, HINGAMP P, QUACKENBUSH J, SHERLOCK G, SPELLMAN P, et al. Minimum information about a microarray experiment (MIAME)-Toward standards for microarray data. Nature Genet. 2001;29(4):365-371.
DRAY S, DUFOUR AB, CHESSEL D. The ade4 package-II: Two table and K-table methods. R News. 2007;7(2):47-52.
EULGEM T. Dissecting the WRKY web of plant defense regulators. PLoS Pathog. 2006;2(11):e126.doi:10.1371/journal.,ppat.0020126.
FIDANTSEF AL, STOUT MJ, THALER JS, DUFFEY SS, BOSTOCK RM. Signal interactions in pathogen and insect attack: Expression of lipoxygenase, proteinase inhibitor II, and pathogenesis-related protein P4 in the tomato, Lycopersicon esculentum. Physiol. Mol Plant Pathol. 1999;54(3-4):97-114.
FRICK UB, SCHALLER A. cDNA microarray analysis of fusicoccin induced changes in gene expression in tomato plants. Planta. 2002;216(1):83-94.
GALLEGLY WE, MARVEL ME. Inheritance of resistance to tomato race 0 of Phytophthora infestans. Phytopathology. 1955;45:103-109.
GIBLY A, BONSHTIEN A, BALAJI V, DEBBIEE P, MARTIN GB, SESSA G. Identification and expression profiling of tomato genes differentially regulated during a resistance response to Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact. 2004;17(11):1212-1222.
GLAZEBROOK J. Genes controlling expression of defense responses in Arabidopsis -2001 status. Curr Opin Plant Biol. 2001;4:301-308.
GU Y, YANG C, THARA VK, ZHOU J, MARTIN GB. The Pti4 gene is regulated by ethylene and salicylic acid and its product is phosphorylated by the Pto kinase. Plant Cell. 2000;12(5):771-785.
HANSSEN IM, VAN ESSE HP, BALLESTER AR, HOGEWONING SW, ORTEGA PARRA N, PAELENAM A, LOEVENS B, BOVY AG, THOMMA BPHJ. Differential tomato transcriptomic responses induced by pepino mosaic virus isolates with differential aggressiveness. Plant Physiology. 2011;156(1):301-318.
HERBERS K, TAKAHATA Y, MELZER M, MOCK HP, HAJIREZAEI M, SONNEWALD U. Regulation of carbohydrate partitioning during the interaction of potato virus Y with tobacco. Mol Plant Pathol. 2000;1(1):51-59.
KOCAL N, SONNEWALD U, SONNEWALD S. Cell Wall-bound invertase limits sucrose export and is involved in symptom development and inhibition of photosynthesis during compatible interaction between tomato and Xanthomonas campestris pv vesicatoria. Plant Physiology. 2008;148(3):1523-1536.
MULLER CH. A revision of the genus Lycopersicon. USDA Misc Publ. 1940;328:1-29.
MYSORE KS, CRASTA OR, TUORI RP, FOLKERTS O, SWIRSKY PB, MARTIN GB. Comprehensive transcript profiling of Pto- and Prfmediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 2002;32(3):299-315.
NIDERMAN T, GENETET I, BRUYERE T, GEES R, STINZI A, LEGRAND M, FRITIG B, MOSINGER E. Pathogenesis-related PR-1 proteins are antifungal. Plant Physiol. 1995;108:17-27.
PERRY KL, FRANCKI RIB. Insect-mediated transmission of mixed and reassorted cucumovirus genomic RNAs. J Gen Virol. 1992;73(Pt 8):2105-2114.
PINZON A, RODRIGUEZ LM, GONZALEZ A, BERNAL A, RESTREPO S. Targeted metabolic reconstruction: a novel approach for the characterization of plant-pathogen interactions. Brief Bioinform. 2011;12(2):151-162.
RESTREPO S, MYERS KL, DEL POZO O, MARTIN GB, HART A, BUELL CR, FRY WE, SMART CD. Gene profiling of a compatible interaction between Phytophthora infestans and Solanum tuberosum suggests a role for carbonic anhydrase. Mol Plant Microbe In. 2005;18(9):913-922.
ROTENBERG D, THOMPPSON TS, GERMANC TL, WILLIS DK. Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J Virol Methods. 2006;138(1-2):49-59.
SCHENK PM, KAZAN K, WILSON I, ANDERSON JP, RICHMOND T, SOMERVILLE SC, MANNERS JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97(21):11655-11660.
SINHA AK, HOFMAN MG, ROMER U, KOCKENBERGER W, WLLINNG L, ROITSCH T. Metabolizable and non-metabolizable sugars activate different signal transduction pathways in tomato. Plant Physiol. 2002;128(4):1480-1489.
SMART CD, WILLMANN MR, MAYTON H, MIZUBUTI ESG, SANDROCK RW, MULDOON AE, FRY WE. Self-fertility in two clonal lineages of Phytophthora infestans. Fungal Genet Biol. 1998; 25(2):134-142.
SMART CD, MYERS KL, RESTREPO S, MARTIN GB, FRY WE. Partial resistance of tomato to Phytophthora infestans is not dependent upon ethylene, jasmonic acid, or salicylic acid signaling pathways. Mol Plant Microbe Interact. 2003;16(2):141-148.
STOUT MJ, FIDANTSEF AL, DUFFEY SS, BOSTOCK RM. Signal interactions in pathogen and insect attack: systemic plant mediated interactions between pathogens and herbivores of the tomato, Lycopersicon esculentum. Physiol Mol Plant Path. 1999;54(3-4):115-130.
TAO Y, XIE Z, CHEN W, GLAZEBROOK J, CHANG HS, HAN B, et al. Quantitative nature of Arabidopsis Responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. The Plant Cell. 2003;15(2):317-330.
TIAN M, WIN J, SONG J, VAN DER HOORN R, VAN DER KNAAP E, KAMOUN SA. Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 2007;143(1):364-377.
TIAN M, BENEDETTI B, KAMOUN SA. Second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic Pathogenesis-related protease P69B of tomato. Plant Physiol. 2005;138(3):1785-1793.
TIAN M, HUITEMA E, DA CUNHA L, TORTO-ALALIBO T, KAMOUN SA. Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem. 2004;279(25):26370-26377.
TUSHER VG, TIBSHIRANI R, CHU G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116-5121.
Cómo citar
APA
ACM
ACS
ABNT
Chicago
Harvard
IEEE
MLA
Turabian
Vancouver
Descargar cita
Visitas a la página del resumen del artículo
Descargas
Licencia
Derechos de autor 2012 Acta Biológica Colombiana

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
1. La aceptación de manuscritos por parte de la revista implicará, además de su edición electrónica de acceso abierto bajo licencia Attribution-NonCommercial-ShareAlike 4.0 (CC BY NC SA), la inclusión y difusión del texto completo a través del repositorio institucional de la Universidad Nacional de Colombia y en todas aquellas bases de datos especializadas que el editor considere adecuadas para su indización con miras a incrementar la visibilidad de la revista.
2. Acta Biológica Colombiana permite a los autores archivar, descargar y compartir, la versión final publicada, así como las versiones pre-print y post-print incluyendo un encabezado con la referencia bibliográfica del articulo publicado.
3. Los autores/as podrán adoptar otros acuerdos de licencia no exclusiva de distribución de la versión de la obra publicada (p. ej.: depositarla en un archivo telemático institucional o publicarla en un volumen monográfico) siempre que se indique la publicación inicial en esta revista.
4. Se permite y recomienda a los autores/as difundir su obra a través de Internet (p. ej.: en archivos institucionales, en su página web o en redes sociales cientificas como Academia, Researchgate; Mendelay) lo cual puede producir intercambios interesantes y aumentar las citas de la obra publicada. (Véase El efecto del acceso abierto).