QTL identification for cassava bacterial blight resistance under natural infection conditions
Identificación de QTL de resistencia a la bacteriosis vascular en yuca bajo condiciones naturales de infección
DOI:
https://doi.org/10.15446/abc.v22n1.57951Palabras clave:
Cassava, molecular marker, QTL, resistance, SNPs, Xanthomonas axonopodis pv. manihotis. (en)marcador molecular, QTL, resistencia, SNPs, Xanthomonas axonopodis pv. manihotis, yuca. (es)
Descargas
Cassava, Manihot esculenta Crantz, represents the main food source for more than one billion people. Cassava’s production is affected by several diseases, one of the most serious is cassava bacterial blight (CBB) caused by Xanthomonas axonopodis pv. manihotis (Xam). A quantitative trait loci (QTL) analysis for CBB resistance was performed under natural infection conditions, using a mapping population of 99 full-sibs genotypes highly segregant and a SNP-based high dense genetic map. The phenotypic evaluation was carried out in Puerto López, Meta, Colombia, during the rainy season in 2015. Both resistant and susceptible transgressive segregants were detected in the mapping population. Through a non-parametric interval mapping analysis, two QTL were detected, explaining 10.9 and 12.6 % of phenotypic variance of resistance to field CBB. After a bioinformatics exploration four genes were identified in the QTL intervals. This work represents a contribution to the elucidation of the molecular bases of quantitative cassava resistance to Xam.
La yuca, Manihot esculenta Crantz, representa la principal fuente de alimento para cerca de 1000 millones de personas. La producción de yuca se ve afectada por diversas enfermedades, una de las más serias es la bacteriosis vascular (CBB) causada por Xanthomonas axonopodis pv. manihotis (Xam). En este estudio se realizó un análisis de loci de rasgos cuantitativos (QTL) para la resistencia a CBB en condiciones naturales de infección, usando una población de mapeo constituida por 99 genotipos de hermanos completos segregantes y un mapa genético altamente denso basado en SNPs. La evaluación fenotípica se llevó a cabo en Puerto López (Meta), Colombia, durante la época de lluvias durante el segundo semestre de 2015. En la población de mapeo fueron detectados individuos con una segregación transgresiva tanto resistentes como susceptibles. A través de un análisis no paramétrico de intervalo simple, se detectaron dos QTL que explican el 10,9 y el 12,6 % de la varianza fenotípica de la resistencia en campo a CBB. Mediante análisis bioinformáticos se identificaron cuatro genes candidatos presentes en los intervalos de los QTL. Este trabajo representa un esfuerzo por dilucidar los mecanismos moleculares implicados en la resistencia de yuca a CBB.
DOI: https://doi.org/10.15446/abc.v22n1.57951
QTL IDENTIFICATION FOR CASSAVA BACTERIAL BLIGHT RESISTANCE UNDER NATURAL INFECTION CONDITIONS
Identificación de QTL de resistencia a la bacteriosis vascular en yuca bajo condiciones naturales de infección
Johana SOTO SEDANO1, Rubén Eduardo MORA MORENO1, Fernando CALLE2, Camilo Ernesto LÓPEZ CARRASCAL1.
1 Departamento de Biología, Universidad Nacional de Colombia, Sede Bogotá. Carrera 45 n.° 26-85, edificio 421, laboratorio 150. Bogotá, Colombia.
2 Unidad de Mejoramiento y Genética de yuca. Centro Internacional de Agricultura Tropical–CIAT. Km 17 Recta Cali-Palmira. Palmira, Colombia.
For correspondence. celopezc@unal.edu.co
Received: 9th June 2016, Returned for revision: 8th November 2016, Accepted: 16th November 2016.
Associate Editor: Geraldo Mäder.
Citation/Citar este artículo como: Soto Sedano J, Mora Moreno RE, Calle F, López Carrascal CE. QTL identification for cassava bacterial blight resistance under natural infection conditions. Acta biol. Colomb. 2017;22(1):19-26. DOI: https://doi.org/10.15446/abc.v22n1.57951
ABSTRACT
Cassava, Manihot esculenta Crantz, represents the main food source for more than one billion people. Cassava's production is affected by several diseases, one of the most serious is cassava bacterial blight (CBB) caused by Xanthomonas axonopodis pv. manihotis (Xam). A quantitative trait loci (QTL) analysis for CBB resistance was performed under natural infection conditions, using a mapping population of 99 full-sibs genotypes highly segregant and a SNP-based high dense genetic map. The phenotypic evaluation was carried out in Puerto López, Meta, Colombia, during the rainy season in 2015. Both resistant and susceptible transgressive segregants were detected in the mapping population. Through a non-parametric interval mapping analysis, two QTL were detected, explaining 10.9 and 12.6 % of phenotypic variance of resistance to field CBB. After a bioinformatics exploration four genes were identified in the QTL intervals. This work represents a contribution to the elucidation of the molecular bases of quantitative cassava resistance to Xam.
Keywords: Cassava, molecular marker, QTL, resistance, SNPs, Xanthomonas axonopodis pv. manihotis.
RESUMEN
La yuca, Manihot esculenta Crantz, representa la principal fuente de alimento para cerca de 1000 millones de personas. La producción de yuca se ve afectada por diversas enfermedades, una de las más serias es la bacteriosis vascular (CBB) causada por Xanthomonas axonopodis pv. manihotis (Xam). En este estudio se realizó un análisis de loci de rasgos cuantitativos (QTL) para la resistencia a CBB en condiciones naturales de infección, usando una población de mapeo constituida por 99 genotipos de hermanos completos segregantes y un mapa genético altamente denso basado en SNPs. La evaluación fenotípica se llevó a cabo en Puerto López (Meta), Colombia, durante la época de lluvias durante el segundo semestre de 2015. En la población de mapeo fueron detectados individuos con una segregación transgresiva tanto resistentes como susceptibles. A través de un análisis no paramétrico de intervalo simple, se detectaron dos QTL que explican el 10,9 y el 12,6 % de la varianza fenotípica de la resistencia en campo a CBB. Mediante análisis bioinformáticos se identificaron cuatro genes candidatos presentes en los intervalos de los QTL. Este trabajo representa un esfuerzo por dilucidar los mecanismos moleculares implicados en la resistencia de yuca a CBB.
Palabras clave: marcador molecular, QTL, resistencia, SNPs, Xanthomonas axonopodis pv. manihotis, yuca.
INTRODUCTION
Cassava, Manihot esculenta Crantz, is a cross-pollinated species and belongs to the Euphorbiaceae family. It is a perennial shrub and its origin is the Amazon Basin (Olsen and Schaal, 1999). The cassava diploid genome is 2n = 36 and has sexual reproduction but for agro-economical purposes farmers use vegetative propagation (Carvalho and War, 2002; Raji et al., 2009). Cassava is one of the most important crops worldwide. It is the third most important source of calories in the tropics, after rice and maize (FAO, 2008). Cassava has been considered essential in protecting food security, especially for developing countries in Africa, Asia and Latin America (FAO, 2008). Due to the high adaptability to drought and acid and poor soils, cassava has been considered as an excellent alternative for an eventual world food crisis (FAO, 2008; FAO, 2013).
Brazil, Thailand, Indonesia, Angola and Ghana are the countries with the largest cassava planted area. Colombia was ranked fifteenth in world cassava production and third in Latin America after Brazil and Paraguay (Aguilera, 2012). In Colombia, Departments such as Bolívar, Córdoba, Sucre, Magdalena and Meta are those with the largest cassava planted area and production. The total production in these Departments was more than 500 thousand tons in 2014 (MINAGRO, 2016).
Cassava, as any other crop, is affected by several diseases produced by virus, fungus, oomycetes and bacteria (FAO, 2013). The most important bacterial disease affecting cassava is Cassava Bacterial Blight (CBB), which is caused by the vascular and foliar pathogen Xanthomonasaxonopodis pv. manihotis (Xam). Recently, Xam was ranked as one of the top 10 most important bacterial phytopathogens (Mansfield et al., 2012). CBB is a devastating disease, generating significant losses, which can reach between 12 and 100 % in infected fields (Lozano, 1986; López and Bernal, 2012). In Colombia, the Xam populations are highly dynamic and diverse (Restrepo et al., 2004; Trujillo et al., 2014).
Conventional breeding strategies have been used to address CBB but with limited success. The most efficient strategy to manage CBB is planting resistant cultivars. However, the knowledge of the molecular mechanisms which governs the resistance in cassava is scarce. Nevertheless, histology and cytochemistry studies of the resistance to CBB shows that callose depositions (Kpémoua, 1996; Sandino et al., 2105), cell wall fortification, lignification and suberization associated with callose deposition and production of flavonoids and polysaccharides are important mechanisms of resistant in response to Xam (Kpémoua et al., 1996). Also, different molecular approaches have conducted to identify resistance and defense genes (López et al., 2003; López et al., 2005).
Resistance to CBB is a quantitative trait, with polygenic and additive inheritance (; Jorge et al., 2000; Jorge et al., 2001). A number of quantitative trait loci (QTL) for resistance to CBB, with major and minor effects as well as stable and unstable have been detected. In 2000, Jorge and coworkers reported twelve QTL explaining 9 % to 27 % of the phenotypic variance. These QTL were detected under greenhouse conditions to five Xam strains (CIO-84, CIO-1, CIO-136, CIO-295 and ORST X-27). Eight novel QTL, explain between 7.2 % and 18.2 % of the resistance, were identified under field conditions of natural disease pressure and during two consecutive crop cycles (Jorge et al., 2001). Nine QTL explaining 16 % to 33 % of the phenotypic variance to four African Xam strains were also reported (Wydra et al., 2004). More recently, two new QTL explaining 62 % and 21 % of the CBB resistance were identified to the Xam strains CIO151 and CIO121 (López et al., 2007).
Undoubtedly the environmental conditions plays a key role in traits governed quantitatively and even more in plant pathogen interactions (Weinig and Schmitt, 2004; Anderson et al., 2014). In fact, several studies had shown that the environment conditions are a key factor in the cassava–Xam interaction. In particular, the humidity favors the dispersion and proliferation of the bacteria and favors the disease (Banito et al., 2000; Wydra and Verdier, 2002; Restrepo, 2004; Banito et al., 2008). Thus, is essential to perform field evaluations with high disease pressure in order to detect genetic determinants involved in CBB resistance under natural conditions where cassava grows.
The objective of this work was to identify novel QTL for CBB field resistance, based on the evaluation of a biparental population of 99 F1 segregating full sib progeny, during a rainy season in 2015 in Meta, Colombia. A bioinformatics research for genes present in the QTL regions was carried out finding some candidate genes.
MATERIALS AND METHODS
Mapping population and field design experiment
The mapping population is a full sib F1 segregating population of 99 individuals obtained by a cross between the Nigerian cultivar TMS30572 and CIAT's (International Center for Tropical Agriculture) elite cultivar CM2177-2 (Fregene, 1997). Due to cassava is a highly heterozygous species, the parental TMS30572 and CM2177-2 are not pure lines, instead are heterozygous, given arise a first progeny (F1) segregating population. This population has been used for several mapping studies (Fregene et al., 1997; Jorge et al., 2000; Jorge et al., 2001; Mba et al., 2001; Lopez et al., 2007), and for the construction of a high-density cassava genetic map (Soto et al., 2015). Each genotype was grown from stem cuttings (stakes) at the research institute "La Libertad" Corpoica, located in Puerto López, Meta, Colombia (4°03'40.3"N 73°27'22.5"W). This region belongs to ecozone 2 (ECZ2): a lowland tropical region in the Colombian eastern plains (Restrepo et al., 1999; Jorge et al., 2001; Trujillo et al., 2014). Ten plants of each parent and each genotype were planted in the field with a density of 1m2, in an area of 1.9 ha under a complete random design. The F1 population was planted in August, 2014. The plants were cultivated according to the agronomical practices employed by the farmers. No control to diseases was conducted. During the evaluation period of the response to natural disease pressure of Xam in Meta, Colombia, in July 2015, corresponding to rainy season (IDEAM, 2015). The maximum and minimal temperatures were 32 °C and 22 °C, respectively, 87 % of relative humidity and a mean precipitation of 400 mm (IDEAM, 2015). Insects or other diseases did not attack the plants, which grew as expected.
The F1 population was planted in August, 2014. The plants were cultivated according to the agronomical practices employed by the farmers. No control to diseases was conducted. During the evaluation period of the response to natural disease pressure of Xam in Meta, Colombia, in July 2015, the maximum and minimal temperatures were 32 °C and 22 °C, respectively, 87 % of relative humidity and a mean precipitation of 400 mm (IDEAM, 2015). Insects or other diseases did not attack the plants, which grew as expected.
Field evaluation of the response to CBB
Under a natural pressure of Xam, the disease severity was scored in ten plants by genotype and parental at ten months after planting, using a rating from 0 to 5, using the symptoms scale described by Jorge et al. (2001). Symptom scale: 1=healthy plant (no symptoms); 2=angular leaf spots; 3=wilting of leaves; 4=dieback of one or several apices; 5=dieback of whole plan. The average of the symptoms at the observation time was calculated for each genotype and taken as a disease index (DI). The DI of each genotype was used for QTL analysis. The transgressive segregants for resistance and susceptibility were also evaluated. The distribution of frequency of the DI was tested for normal distribution by the Shapiro-Wilk test. An analysis of variance (ANOVA) and its non-parametric test (Kruskal-Wallis) were also performed.
QTL mapping
Interval Mapping (IM) analysis with the "np" model was used for QTL detection through R/qtl V1.37-11 (Broman, 2015). The high dense genetic map of cassava with a density or distance between markers of 1,26 cM (Soto et al., 2015) was employed plus a set of 2,236 GBS-SNP markers with unknown genetic position but with known physical position in the current cassava genome v6.1. To declare the presence of a QTL a LOD score higher than 2.5 was used as criteria. The QTL interval was established by a LOD decrease of 0.5 from the marker peak position. Phenotypic variation explained by each QTL was determined with the R package calc.Rsq. Physical positions of the genes identified within the QTL intervals (candidate genes) were established based on the SNP-based genetic map. A BLAST (Basic Local Alignment Search Tool) analysis were performed to the current cassava genome (v6.1) for genes identified within the QTLs intervals.
RESULTS
Field evaluation of the response to cassava bacterial blight
At the end of the productive cycle (ten months after planting) and before harvest the roots, the cassava plants were scored for the presence of symptoms. Five genotypes (5 %) were symptomless, while 94 genotypes (95 %) exhibit at least one of the typical symptoms related to CBB, being the angular leaf spots the most common. Taking into account these observations it is possible to conclude that CBB was present in the field and in consequence it was possible to evaluate the differential responses among the genotypes. The plants were categorized according to the presence of symptoms using a field scale previously established (Jorge et al., 2001) and a disease index (DI) was calculated for each genotype as the number of symptomatic plants over the total of evaluated plants. The DI in the mapping population did not exhibit a normal distribution, (p<0.05) (Fig. 1). However, the Kruskal-Wallis test showed significant differences (p<0.05) for the DI values obtained for the genotypes tested, showing that the response to CBB is genotype-dependent (Supplementary data). Both parents exhibited DIs statistically different (significant p<0.05), indicating contrasting responses to CBB (Supplementary data). The resistant parental TMS30572 had a DI value of 0.2 while the DI for the susceptible parent was 0.6. The DI in the mapping population ranged from 0 to 2, with a mean of 0.75 and a standard deviation of 0.45. Most DIs values were found near the average of the sample (34 genotypes with DI=1) and very few values near the upper (four genotypes with DI >1.8) and lower extremes (five genotypes with DI=0) (Fig. 1). From the 99 genotypes evaluated, 38 have equal or lower DI values than 0.2 (DI value of the resistant parent TMS30572), these were considered as resistant genotypes. On the other hand, 61 genotypes exhibited equal or higher DI values than 0.6 (DI value for the susceptible parent CM772-14) and those were considered as susceptible.
Transgressive segregants with DIs higher or lower than the parents were identified in the mapping population. The total trasngressive segregants were eight for DIs lower than 0.2 (ID value of the parent TMS30572) and 60 higher than 0.6 (ID value of the parent CM2177-2). The genotypes g52 and g135 were the extreme genotypes for susceptibility with ID values of 1.83 and 2, respectively. For resistance, the extreme genotypes were g23, g89, g92, g93 and g104, which did not exhibit any symptom related to CBB (Supplementary data).
QTL mapping
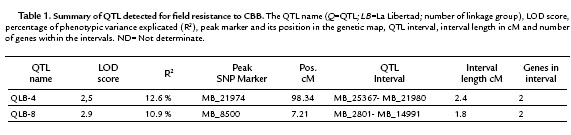
Due to the ID values in the mapping population did not exhibit a normal distribution, a non-parametric QTL interval mapping approach using the model "np" of R/qtl, was applied. Based on the phenotyping evaluation of the response to CBB in the F1 population and the previous cassava genetic map developed (Soto et al., 2015). This analysis allowed the identification of two QTL explaining CBB field resistance. These QTL were located in linkage groups 4 and 8 with LOD 2.5 and 2.9 respectively (Fig. 2). The QTL in the linkage group 4 was named as QLB-4 and explains the 12.6% of the field resistance to CBB. It covers an interval length of 2.4 cM. The interval flanking markers of QLB-4 were MB_21980 and MB_25367. The physical distance from the peak (MB_21974) to the flanking marker MB_21980 was 317bp. While the distance from the peak marker to the flanking marker MB_24367 could not be established due to these two markers belong to different scaffolds in the cassava genome v4.1.
The second QTL was located on the linkage group 8, explained 10.9 % of CBB resistance and was named as QLB-8. The QLB-8 covers an interval of 1.8 cM, whit a peak marker matching to the SNP MB_8500 and interval flanking markers MB_2801 and MB_14991 (Table 1). The physical distance from the peak marker to the flanking markers could not be established due to the two flanking markers belonging to different scaffolds in the cassava genome v4.1.
For each QTL interval, the corresponding genomic regions were searched for the presence of coding regions based on the new cassava genome version v6.1 (phytozome.com). The positions of the SNPs markers MB_21974 (peak marker) and MB_21980 (flanking marker) of QLB-4 match with the gene Manes.07G062100. According to PFAM (Protein Family) the annotation, this gene coded for a protein related to the vacuolar-sorting receptor 3. The position of the other flanking marker of QLB-4 (MB_25367) matches with the gene Manes.07G053100 which following the PFAM annotation corresponds to a serine protease carboxypeptidase. While in the interval of QLB-8 two genes were detected: Manes.S010100a and Manes.03G002800, which code for a C2HC zinc finger-containing protein and for a core-2/i-branching beta-1, 6-n-acetylglucosaminyltransferase protein, respectively.
DISCUSSION
The present study evaluated the phenotypic response of 99 full-sib segregant genotypes to CBB in field during the rainy season at Meta (Eastern Plains), one of the most productive areas of cassava in Colombia (Agronet-MINAGRO, 2016). The Colombian Eastern plains belong to the ECZ2 which is characterized by savannas of acid soils, with mean temperature of 26.1 °C and mean precipitation of 400 mm per month (Jorge et al., 2001; Ospina et al., 2002; Restrepo et al., 2004). This ECZ2 has been described as an area with one of the highest incidence of CBB in Colombia (Jorge, 2001). Several studies have shown that Xam populations present ecozone-differentiation as well as pathogenic specialization to the local adapted cassava material (Restrepo and Verdier 1997; Restrepo, 1999). Thus, the QTL reported here could be useful for further breeding strategies whose interest will be developing new CBB-resistant cassava varieties highly adapted to this ECZ. It will be important to carry out studies on the pathogen in this area in order to dissect the current status of the presence of different Xam strains and its dynamics. The last information available on Xam in this particular ECZ was obtained almost two decades ago (Restrepo, 1999).
An adequate high disease pressure was observed during the field evaluation. Differences in the severity of the disease between parental genotypes, as well as differences within the individuals of the mapping population, could be detected. Based on the phenotype data obtained it was possible the identification of two QTL explaining 10.9 and 12.6 % of cassava resistance to Xam. In each of these QTL regions were found two coding genes, representing novel candidate genes for CBB resistance.
A higher number of susceptible genotypes (61.6 %) compared to the susceptible genotypes were identified in the mapping population. Due to the phenotypic evaluation was performed during a rainy year, it is expected that the high humidity had contributed to this finding. This is consistent with several studies showing that high humidity favored the development and speed of symptoms as well Xam growing and dispersion (Leu, 1978; Banito et al., 2000; Banito et al., 2001; Wydra and Verdier, 2002; Restrepo, 2004).
Both, resistant and susceptible transgressive segregants were identified in the phenotypic evaluation of the mapping population. Ten resistant transgressive genotypes were identified. This type of segregation has been described for several crops (Akinwale et al., 2010; Whankaew et al., 2011; Thanyasiriwat et al., 2013; Njenga et al., 2014), as well for cassava to CBB resistance in the same mapping population used in this study (Jorge et al., 2000; Jorge et al., 2001). The finding of these segregants suggests the action of additive and dominant genes for CBB resistance in the TMS30571 x CM2177-2 cross. The transgressive genotypes and especially the extreme resistant individuals g23, g89, g92, g93 and g104, became an important source of CBB resistance to be employed in different cassava breeding programs.
In this study two QTL were identified. A previous QTL detection study for CBB conducted also at ECZ2 (Jorge et al., 2001) revealed the presence of eight QTL from which six were stable. On greenhouse and controlled conditions with particular Xam strains more than twenty QTL have been identified (Jorge et al., 2000; Wydra et al., 2004; Lopez et al., 2007). Several of these QTL were strain-specific QTL. The identification of several resistance QTLs have been achieved through QTL mapping in important pathosystems, some of the most recently reports include QTL the resistance to virus diseases affecting maize (Wang et al., 2016), leaf rust in wheat (Soriano and Royo, 2015) and Xanthomonas oryzae pv. oryzae in rice (Dejedatin et al., 2016) within others.
In order to evaluate the stability of the QTL reported here, it will be necessary to perform additional field evaluations during different years and dry seasons. Another important aspect will be to determinate the Xam strain (s) present on the infected plants to know if these QTL are strain specific.
The two QTL identified in this study cover a genetic region of 4.2 cM, 2.4 cM for QLB-4 and 1.8 cM for QLB-8, respectively. The QTL cover a short interval length given the high marker density exhibited for this genetic map (Soto et al., 2015). In addition, this genetic map was anchored to the cassava genome which allowed the identification of the genes present in the QTL intervals. Even though some associations of candidate genes with QTL have been reported (Faris et al., 1999; Ramalingam et al., 2003; Liu et al., 2004), these types of studies are scarce. Here it was possible to identify four candidate genes in a relative short interval length. These genes are represented by the molecular markers located within the QTL interval. Due to those interval markers belong to different cassava genome scaffolds a physical map region was not established, in consequence, it is possible that more than four genes were located within the physical interval. Nevertheless, the presence of two genes in each of these QTL will facilitate the number of genes to be functionally validated.
The functional annotation of the four genes present within the QTL intervals are not directly related to known plant immunity related genes. Other studies have reported the presence of genes coding for proteins related in plant immunity process as pathogen perception or in signal pathways (Ramalingam et al., 2003; St. Clair, 2010; Lopez, 2011). However, some studies have established that the typical immunity-related genes are only a small part of the whole genes related to plant resistance (Corwin et al., 2016). Recently, several genes have been cloned from QTL and none of them correspond to classical R genes, but have different functions not directly related with pathogen recognition or defense (Poland et al., 2009; Bryant et al., 2014; Roux et al., 2014). Thus, the four genes here detected become new genetic factors that may be playing an important role in CBB resistance. The functional validation of these genes should be addressed in order to deepen the understanding of the cassava response to Xam.
CONCLUSIONS
In this study, the phenotypic evaluation of the response of 99 full-sib segregant genotypes to Xam infection in field conditions in Meta, Colombia, allowed the detection of two novel QTL associated to CBB resistance. These QTL explained 10.9 and 12.6 % of the field resistance to the disease. Four genes were identified in the QTL intervals. The genes coding for a protein related to the vacuolar-sorting receptor, a serine protease carboxypeptidase, a C2HC zinc finger-containing protein and for a core-2/i-branching beta-1,6-n-acetylglucosaminyltransferase protein. These genes become novel genetic factors which can be playing a role in plant resistance to Xam. The QTL and the genes involved in CBB field resistance detected in this study can contribute in local marker assisted-breeding strategies and further map-based studies focus on dissect the major responsible genes governing CBB resistance.
ACKNOWLEDGMENTS
We want to thank Universidad Nacional de Colombia and COLCIENCIAS for the PhD scholarship call 528 of the author Johana Soto. We would like to extend our gratitude to the "Unidad de Mejoramiento y Genética"- CIAT International Center for Tropical Agriculture (CIAT) for enabling the plant material and to the staff of Corpoica La libertad for the support during the field experiments. Supplementary data are available at Online version.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
Agronet, MINAGRO. Ministerio de Agricultura y Desarrollo Rural. República de Colombia. 2016. Available in: http://agronet.gov.co. Accessed 10-05-2016.
Aguilera. La yuca en el Caribe colombiano: De cultivo ancestral a agroindustrial. 2012. Available in: http://www.banrep.gov.co/documentos/publicaciones/regional/documentos/DTSER-158.pdf.
Anderson J, Wagner M, Rushworth C, Prasad K, Mitchell-Olds T. The evolution of quantitative traits in complex environments. Heredity, 2014;112 (1):4-12. Doi: 10.1038/hdy.2013.33.
Akinwale M, Aladesanwa R, Akinyele B, Dixon A, Odiyi A. Inheritance of ß-carotene in cassava (Manihot esculenta crantz). Int J Genet Mol Biol. 2010;2(10):198-201.
Banito A, Kpémoua K, Wydra K, Rudolph K. Bacterial blight of cassava in Togo: its importance, the virulence of the pathogen and the resistance of varieties. Plant Patho Bact. 2001; Chapter:259-264.
Banito A, Kpemoua K, Wydra K. Expression of resistance and tolerance of cassava genotypes to bacterial blight determined by genotype x environment interactions. J Plant Dise Prot. 2008;115(4):152-161. Doi: 10.1007/BF03356261.
Broman K. R/qtlcharts: interactive graphics for quantitative trait locus mapping. Genetics. 2015;199(2):359-361. Doi: 10.1534/genetics.114.172742.
Bryant R, McGrann R, Mitchell R, Schoonbeek J, Boyd A, Uauy C, Ridout C. A change in temperature modulates defence to yellow (stripe) rust in wheat line UC1041 independently of resistance gene Yr36. BMC Plant Biol. 2014;14(1):1. Doi: 10.1186/1471-2229-14-10.
Carvalho R, Guerra M. Cytogenetics of Manihot esculenta Crantz (cassava) and eight related species. Hereditas. 2002;136:159-168. Doi: 10.1034/j.1601-5223.2002.1360212.x.
Corwin J, Copeland D, Feusier J, Subedy A, Eshbaugh R, Palmer C, Kliebenstein D. The quantitative basis of the Arabidopsis innate immune system to endemic pathogens depends on pathogen genetics. PLoS Genet. 2016;12(2):e1005789. Doi: 10.1371/journal.pgen.1005789.
Djedatin G, Ndjiondjop M, Sanni A, Lorieux M, Verdier V, Ghesquiere A. Identification of novel major and minor QTLs associated with Xanthomonas oryzae pv. oryzae (African strains) resistance in rice (Oryza sativa L.). Rice. 2016;9(1):1-10. Doi: 10.1186/s12284-016-0090-9.
FAO. Oficina de prensa. Yuca para la seguridad alimentaria y energética. 2008. Available in: http://www.fao.org/newsroom/es/news/2008/1000899/index.html. Accessed 13-05-2016.
FAO. Panorama de la seguridad alimentaria y nutricional de América Latina y el Caribe. Santiago: Oficina Regional para América Latina y el Caribe de FAO; 2013. p. 48.
Faris J, Liu D, Chen P, Gill B. Candidate gene analysis of quantitative disease resistance in wheat. Theor Appl Genet. 1999;98:219-225. Doi: 10.1007/s001220051061.
Fregene M., Angel F, Gómez R, Rodríguez F, Chavarriaga P, Roca W, Bonierbale M. A molecular genetic map of cassava (Manihot esculenta Crantz). Theor Appl Genet. 1997;95(3):431-441. Doi: 10.1007/s001220050580.
IDEAM (Instituto de Hidrología, Meteorología y Estudios Ambientales). Available in: www.ideam.gov.co Sistema de información ambiental, registros estacionales. Estación Puerto Lopez, (Meta). 2015. Accessed 10-05-2016.
Kpémoua K, Boher B, Nicole M., Calatayud P, Geiger J. Cytochemistry of defense responses in cassava infected by Xanthomonas campestris pv. manihotis. Can J Microbiol. 1996;42(11):1131-1143. Doi: 10.1139/m96-145.
Jorge V, Fregene M, Duque M, Bonierbale M, Tohme J, Verdier V. Genetic mapping of resistance to bacterial blight disease in cassava (Manihot esculenta Crantz). Theor Appl Genet. 2000;101(5-6):865-872.
Jorge V, Fregene M, Vélez C, Duque M, Tohme J, Verdier V. QTL analysis of field resistance to Xanthomonas axonopodis pv. manihotis in cassava. Theor Appl Genet. 2001;102(4):564-571. Doi: 10.1007/s001220051683.
Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Toth I. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13(6):614-629. Doi: 10.1111/j.1364-3703.2012.00804.x.
Mba R, Stephenson P, Edwards K, Melzer S, Nkumbira J, Gullberg U and Fregene M. Simple sequence repeat (SSR) markers survey of the cassava (Manihot esculenta Crantz) genome: towards an SSR-based molecular genetic map of cassava. Theor Appl Genet. 2001;102(1):21-31. Doi: 10.1007/s001220051614.
Njenga P, Edema R, Kamau J. Combining ability for beta-carotene and important quantitative traits in a cassava F1 population.J. Plant Breed. Crop Sci. 2014;6(2):24-30. Doi: 10.5897/JPBCS12.069.
Lopez C, Zuluaga A, Cooke R, Delseny M, Tohme J, Verdier V. Isolation of resistance gene candidates (RGCs) and characterization of an RGC cluster in cassava. Mol Genet Genomics. 2003;269(5):658-671. Doi: 10.1007/s00438-003-0868-5.
Lopez C, Soto M, Restrepo S, Piégu B, Cooke R, Delseny M, Verdier V. Gene expression profile in response to Xanthomonas axonopodis pv. manihotis infection in cassava using a cDNA microarray. Mol Plant Pathol. 2005;57(3):393-410. Doi: 10.1007/s11103-004-7819-3.
Lopez C, Quesada L, Bohorquez A, Duque M, Vargas J, Tohme J, et al. Mapping EST-derived SSRs and ESTs involved in resistance to bacterial blight in Manihot esculenta. Genome. 2007;50:1078-1088. Doi: 10.1139/G07-087.
Lopez C. Descifrando las bases moleculares de la resistencia cuantitativa. Acta biol Colomb. 2011;16(2):3-16.
Lozano C. Cassava Bacterial Blight: A manageable disease. Plant Dis. 1986;70(12):1989-1993.
Olsen K, Schaal B. Evidence on the origin of cassava: phylogeography of Manihot esculenta. Proc Natl Acad Sci U S A. 1999;96(10):5586-5591. Doi: 10.1073/pnas.96.10.5586.
Ospina B, Ceballos H. La yuca en el tercer Milenio: Sistemas Modernos de producción, procesamiento, utilización y comercialización. 2002; 327. CIAT.
Poland J, Balint-Kurti P, Wisser R, Pratt C, Nelson J. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 2009;14(1):21-29. Doi: 10.1016/j.tplants.2008.10.006.
Raji A, Anderson J, Kolade O, Ugwu C, Dixon A, Ingelbrecht I. Gene-based microsatellites for cassava (Manihot esculenta Crantz): prevalence, polymorphisms, and cross-taxa utility. BMC Plant Biol.2009; 9(1):118. Doi: 10.1186/1471-2229-9-118.
Ramalingam J, Vera C, Kukreja K, Chittoor J, Wu J, Lee S, Leach J. Candidate defense genes from rice, barley, and maize and their association with qualitative and quantitative resistance in rice. Mol. Plant Microbe Interact. 2003;16(1):14-24. Doi: 10.1094/MPMI.2003.16.1.14.
Restrepo S, Verdier V. Geographical differentiation of the population of Xanthomonas axonopodis pv. manihotis in Colombia. Appl Environ Microbiol. 1997;63:4427-4434.
Restrepo S. Etude de la structure des populations de Xanthomonas axonopodis pv. manihotis en Colombie. 1999. PhD thesis, University of Paris VI, France.
Restrepo S, Velez C, Duque M, Verdier V. Genetic structure and population dynamics of Xanthomonas axonopodis pv. manihotis in Colombia from 1995 to 1999. Appl Environ Microbiol. 2004;70(1):255-261. Doi: 0.1128/AEM.70.1.255-261.
Roux F, Voisin D, Badet T, Balagué C, Barlet X, Huard Chauveau C, Raffaele S. Resistance to phytopathogens e tutti quanti: placing plant quantitative disease resistance on the map. Mol Plant Pathol. 2014;15(5):427-432. Doi: 10.1111/mpp.12138.
Sandino T, López-Kleine L, López C, Marquínez X. Characterization of the morphological response of susceptible and resistant varieties of cassava (Manihot esculenta Crantz) to vascular bacterial blight caused by Xanthomonas axonopodis pv manihotis. Summa Phytopathol. 2015;41(2):94-100. Doi: 10.1590/0100-5405/2031.
Soriano J, Royo C. Dissecting the genetic architecture of leaf rust resistance in wheat by QTL meta-analysis. Phytopathology. 2015;105:1585-1593. Doi: 10.1094/PHYTO-05-15-0130-R.
Soto J, Ortiz J, Perlaza-Jiménez L, Vásquez A, Lopez-Lavalle L, Mathew B, et al. A genetic map of cassava (Manihot esculenta Crantz) with integrated physical mapping of immunity-related genes. BMC Genomics. 2015;16;(190). Doi: 10.1186/s12864-015-1397-4.
St. Clair D. Quantitative disease resistance and quantitative resistance loci in breeding. Annu Rev Phytopatho. 2010;(48):247-268.
Thanyasiriwat T, Sraphete S, Whankaew S, Triwitayakorn K. Quantitative trait loci and candidate genes associated with starch pasting viscosity characteristics in cassava (Manihot esculenta Crantz). Plant Bio. 2013;16(1). Doi: 10.1111/plb.12022.
Trujillo C, Ochoa J, Mideros M, Restrepo S, López C, Bernal A. A complex population structure of the Cassava Pathogen Xanthomonas axonopodis pv. manihotis in recent years in the Caribbean Region of Colombia. Microbial ecology. 2014;68(1): 155-167. Doi: 10.1007/s00248-014-0411-8.
Wang Y, Xu J, Deng D, Ding H, Bian Y, Yin Z, Wu Y, Zhou B, Zhao Y. A comprehensive meta-analysis of plant morphology, yield, stay-green, and virus disease resistance QTL in maize (Zea mays L.). Planta. 2016; 243:459-471. Doi: 10.1007/s00425-015-2419-9.
Whankaew S, Poopear S, Kanjanawattanawong S, Tangphatsornruang S, Boonseng O, Lightfoot D, Triwitayakorn K. A genome scan for quantitative trait loci affecting cyanogenic potential of cassava root in an outbred population. BMC genomics. 2011;12(1):1. Doi: 10.1186/1471-2164-12-266.
Weinig, C, Schmitt J. Environmental effects on the expression of quantitative trait loci and implications for phenotypic evolution. Bioscience. 2004;54(7):627-635. Doi: 10.1641/0006-3568.
Wydra K, Verdier V. Occurrence of cassava diseases in relation to environmental, agronomic and plant characteristics. Agric Ecosyst Environ. 2002;93(1): 211-226.
Wydra K, Zinsou V, Jorge V and Verdier V. Identification of pathotypes of Xanthomonas axonopodis pv. manihotis in Africa and detection of quantitative trait loci and markers for resistance to bacterial blight of cassava. Phytopathol. 2004;94(10):1084-1093. Doi: 10.1094/PHYTO.2004.94.10.1084.
Soto Sedano J, Mora Moreno RE, Calle F, López Carrascal CE.
Referencias
Agronet, MINAGRO. Ministerio de Agricultura y Desarrollo Rural. República de Colombia. 2016. Available in: http://agronet.gov.co. Accessed 10-05-2016.
Aguilera. La yuca en el Caribe colombiano: De cultivo ancestral a agroindustrial. 2012. Available in: http://www.banrep.gov.co/documentos/publicaciones/regional/documentos/ DTSER-158.pdf
Anderson J, Wagner M, Rushworth C, Prasad K, Mitchell-Olds T. The evolution of quantitative traits in complex environments. Heredity, 2014;112 (1):4-12. Doi: 10.1038/hdy.2013.33.
Akinwale M, Aladesanwa R, Akinyele B, Dixon A, Odiyi A. Inheritance of ß-carotene in cassava (Manihot esculenta crantz). Int J Genet Mol Biol. 2010;2(10):198-201.
Banito A, Kpémoua K, Wydra K, Rudolph K. Bacterial blight of cassava in Togo: its importance, the virulence of the pathogen and the resistance of varieties. Plant Patho Bact. 2001; Chapter:259-264.
Banito A, Kpemoua K, Wydra K. Expression of resistance and tolerance of cassava genotypes to bacterial blight determined by genotype x environment interactions. J Plant Dise Prot. 2008;115(4):152-161. Doi: 10.1007/BF03356261.
Broman K. R/qtlcharts: interactive graphics for quantitative trait locus mapping. Genetics. 2015;199(2):359-361. Doi: 10.1534/genetics.114.172742.
Bryant R, McGrann R, Mitchell R, Schoonbeek J, Boyd A, Uauy C, Ridout C. A change in temperature modulates defence to yellow (stripe) rust in wheat line UC1041 independently of resistance gene Yr36. BMC Plant Biol. 2014;14(1):1. Doi: 10.1186/1471-2229-14-10.
Carvalho R, Guerra M. Cytogenetics of Manihot esculenta Crantz (cassava) and eight related species. Hereditas. 2002;136:159-168. Doi: 10.1034/j.1601-5223.2002.1360212.x.
Corwin J, Copeland D, Feusier J, Subedy A, Eshbaugh R, Palmer C, Kliebenstein D. The quantitative basis of the Arabidopsis innate immune system to endemic pathogens depends on pathogen genetics. PLoS Genet. 2016;12(2):e1005789. Doi: 10.1371/journal.pgen.1005789.
Djedatin G, Ndjiondjop M, Sanni A, Lorieux M, Verdier V, Ghesquiere A. Identification of novel major and minor QTLs associated with Xanthomonas oryzae pv. oryzae (African strains) resistance in rice (Oryza sativa L.). Rice. 2016;9(1):1-10. Doi: 10.1186/s12284-016-0090-9.
FAO. Oficina de prensa. Yuca para la seguridad alimentaria y energética. 2008. Available in: http://www.fao.org/newsroom/es/news/2008/1000899/index.html. Accessed 13-05-2016.
FAO. Panorama de la seguridad alimentaria y nutricional de América Latina y el Caribe. Santiago: Oficina Regional para América Latina y el Caribe de FAO; 2013. p. 48.
Faris J, Liu D, Chen P, Gill B. Candidate gene analysis of quantitative disease resistance in wheat. Theor Appl Genet. 1999;98:219-225. Doi: 10.1007/s001220051061.
Fregene M., Angel F, Gómez R, Rodríguez F, Chavarriaga P, Roca W, Bonierbale M. A molecular genetic map of cassava (Manihot esculenta Crantz). Theor Appl Genet. 1997;95(3):431-441. Doi: 10.1007/s001220050580.
IDEAM (Instituto de Hidrología, Meteorología y Estudios Ambientales). Available in: www.ideam.gov.co Sistema de información ambiental, registros estacionales. Estación Puerto Lopez, (Meta). 2015. Accessed 10-05-2016.
Kpémoua K, Boher B, Nicole M., Calatayud P, Geiger J. Cytochemistry of defense responses in cassava infected by Xanthomonas campestris pv. manihotis. Can J Microbiol. 1996;42(11):1131-1143. Doi: 10.1139/m96-145.
Jorge V, Fregene M, Duque M, Bonierbale M, Tohme J, Verdier V. Genetic mapping of resistance to bacterial blight disease in cassava (Manihot esculenta Crantz). Theor Appl Genet. 2000;101(5-6):865-872. Doi:
Jorge V, Fregene M, Vélez C, Duque M, Tohme J, Verdier V. QTL analysis of field resistance to Xanthomonas axonopodis pv. manihotis in cassava. Theor Appl Genet. 2001;102(4):564-571. Doi: 10.1007/s001220051683.
Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Toth I. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13(6):614-629. Doi:10.1111/j.1364-3703.2012.00804.x
Mba R, Stephenson P, Edwards K, Melzer S, Nkumbira J, Gullberg U and Fregene M. Simple sequence repeat (SSR) markers survey of the cassava (Manihot esculenta Crantz) genome: towards an SSR-based molecular genetic map of cassava. Theor Appl Genet. 2001;102(1):21-31. Doi: 10.1007/s001220051614.
Njenga P, Edema R, Kamau J. Combining ability for beta-carotene and important quantitative traits in a cassava F1 population.J. Plant Breed. Crop Sci. 2014;6(2):24-30. Doi:10.5897/JPBCS12.069.
Lopez C, Zuluaga A, Cooke R, Delseny M, Tohme J, Verdier V. Isolation of resistance gene candidates (RGCs) and characterization of an RGC cluster in cassava. Mol Genet Genomics. 2003;269(5):658-671. Doi: 10.1007/s00438-003-0868-5.
Lopez C, Soto M, Restrepo S, Piégu B, Cooke R, Delseny M, Verdier V. Gene expression profile in response to Xanthomonas axonopodis pv. manihotis infection in cassava using a cDNA microarray. Mol Plant Pathol. 2005;57(3):393-410. Doi: 10.1007/s11103-004-7819-3.
Lopez C, Quesada L, Bohorquez A, Duque M, Vargas J, Tohme J, et al. Mapping EST-derived SSRs and ESTs involved in resistance to bacterial blight in Manihot esculenta. Genome. 2007;50:1078-1088. Doi:10.1139/G07-087
Lopez C. Descifrando las bases moleculares de la resistencia cuantitativa. Acta biol Colomb. 2011;16(2):3-16.
Lozano C. Cassava Bacterial Blight: A manageable disease. Plant Dis. 1986;70(12):1989-1993.
Olsen K, Schaal B. Evidence on the origin of cassava: phylogeography of Manihot esculenta. Proc Natl Acad Sci U S A. 1999;96(10):5586-5591. Doi: 10.1073/pnas.96.10.5586.
Ospina B, Ceballos H. La yuca en el tercer Milenio: Sistemas Modernos de producción, procesamiento, utilización y comercialización. 2002; 327. CIAT.
Poland J, Balint-Kurti P, Wisser R, Pratt C, Nelson J. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 2009;14(1):21-29. Doi: 10.1016/j.tplants.2008.10.006.Raji A, Anderson J, Kolade O, Ugwu C, Dixon A, Ingelbrecht I. Gene-based microsatellites for cassava (Manihot esculenta Crantz): prevalence, polymorphisms, and cross-taxa utility. BMC Plant Biol.2009; 9(1):118. Doi: 10.1186/1471-2229-9-118.
Ramalingam J, Vera C, Kukreja K, Chittoor J, Wu J, Lee S, Leach J. Candidate defense genes from rice, barley, and maize and their association with qualitative and quantitative resistance in rice. Mol. Plant Microbe Interact. 2003;16(1):14-24. Doi: 10.1094/MPMI.2003.16.1.14.
Restrepo S, Verdier V. Geographical differentiation of the population of Xanthomonas axonopodis pv. manihotis in Colombia. Appl Environ Microbiol. 1997;63:4427-4434.
Restrepo S. Etude de la structure des populations de Xanthomonas axonopodis pv. manihotis en Colombie. 1999. PhD thesis, University of Paris VI, France.
Restrepo S, Velez C, Duque M, Verdier V. Genetic structure and population dynamics of Xanthomonas axonopodis pv. manihotis in Colombia from 1995 to 1999. Appl Environ Microbiol. 2004;70(1):255-261. Doi: 0.1128/AEM.70.1.255-261.
Roux F, Voisin D, Badet T, Balagué C, Barlet X, Huard‐Chauveau C, Raffaele S. Resistance to phytopathogens e tutti quanti: placing plant quantitative disease resistance on the map. Mol Plant Pathol. 2014;15(5):427-432. Doi: 10.1111/mpp.12138.
Sandino T, López-Kleine L, López C, Marquínez X. Characterization of the morphological response of susceptible and resistant varieties of cassava (Manihot esculenta Crantz) to vascular bacterial blight caused by Xanthomonas axonopodis pv manihotis. Summa Phytopathol. 2015;41(2):94-100. Doi: 10.1590/0100-5405/2031.
Soriano J, Royo C. Dissecting the genetic architecture of leaf rust resistance in wheat by QTL meta-analysis. Phytopathology. 2015;105:1585-1593. Doi: 10.1094/PHYTO-05-15-0130-R,
Soto J, Ortiz J, Perlaza-Jiménez L, Vásquez A, Lopez-Lavalle L, Mathew B, et al. A genetic map of cassava (Manihot esculenta Crantz) with integrated physical mapping of immunity-related genes. BMC Genomics. 2015;16;(190). Doi: 10.1186/s12864-015-1397-4.
St. Clair D. Quantitative disease resistance and quantitative resistance loci in breeding. Annu Rev Phytopatho. 2010;(48):247-268.
Thanyasiriwat T, Sraphete S, Whankaew S, Triwitayakorn K. Quantitative trait loci and candidate genes associated with starch pasting viscosity characteristics in cassava (Manihot esculenta Crantz). Plant Bio. 2013;16(1). Doi: 10.1111/plb.12022.
Trujillo C, Ochoa J, Mideros M, Restrepo S, López C, Bernal A. A complex population structure of the Cassava Pathogen Xanthomonas axonopodis pv. manihotis in recent years in the Caribbean Region of Colombia. Microbial ecology. 2014;68(1): 155-167. Doi: 10.1007/s00248-014-0411-8.
Wang Y, Xu J, Deng D, Ding H, Bian Y, Yin Z, Wu Y, Zhou B, Zhao Y. A comprehensive meta-analysis of plant morphology, yield, stay-green, and virus disease resistance QTL in maize (Zea mays L.). Planta. 2016; 243:459-471. Doi: 10.1007/s00425-015-2419-9.
Whankaew S, Poopear S, Kanjanawattanawong S, Tangphatsornruang S, Boonseng O, Lightfoot D, Triwitayakorn K. A genome scan for quantitative trait loci affecting cyanogenic potential of cassava root in an outbred population. BMC genomics. 2011;12(1):1. Doi: 10.1186/1471-2164-12-266.
Weinig, C, Schmitt J. Environmental effects on the expression of quantitative trait loci and implications for phenotypic evolution. Bioscience. 2004;54(7):627-635. Doi: 10.1641/0006-3568.
Wydra K, Verdier V. Occurrence of cassava diseases in relation to environmental, agronomic and plant characteristics. Agric Ecosyst Environ. 2002;93(1): 211-226.
Wydra K, Zinsou V, Jorge V and Verdier V. Identification of pathotypes of Xanthomonas axonopodis pv. manihotis in Africa and detection of quantitative trait loci and markers for resistance to bacterial blight of cassava. Phytopathol. 2004;94(10):1084-1093. Doi: 10.1094/PHYTO.2004.94.10.1084.
Cómo citar
APA
ACM
ACS
ABNT
Chicago
Harvard
IEEE
MLA
Turabian
Vancouver
Descargar cita
CrossRef Cited-by
1. U. N. Ikeogu, I. C. Okwuonu, N. R. Okereke, L. C. Jibuwa, C. Nwadili, S. P. Abah, L. A. Nwachukwu, I. C. Nnaji, C. K. Nkere, J. T. Onyeka, C. N. Egesi. (2022). Genomic Designing for Biotic Stress Resistant Technical Crops. , p.1. https://doi.org/10.1007/978-3-031-09293-0_1.
2. André Antoine Fanou, Valerien Amégnikin Zinsou, Kerstin Wydra. (2018). Cassava. https://doi.org/10.5772/intechopen.71527.
3. Edwige Gaby Nkouaya Mbanjo, Ismail Yusuf Rabbi, Morag Elizabeth Ferguson, Siraj Ismail Kayondo, Ng Hwa Eng, Leena Tripathi, Peter Kulakow, Chiedozie Egesi. (2021). Technological Innovations for Improving Cassava Production in Sub-Saharan Africa. Frontiers in Genetics, 11 https://doi.org/10.3389/fgene.2020.623736.
4. Piengtawan Tappiban, Supajit Sraphet, Nattaya Srisawad, Duncan R Smith, Kanokporn Triwitayakorn. (2018). Identification and expression of genes in response to cassava bacterial blight infection. Journal of Applied Genetics, 59(4), p.391. https://doi.org/10.1007/s13353-018-0457-2.
Dimensions
PlumX
Visitas a la página del resumen del artículo
Descargas
Licencia
Derechos de autor 2017 Acta Biológica Colombiana

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
1. La aceptación de manuscritos por parte de la revista implicará, además de su edición electrónica de acceso abierto bajo licencia Attribution-NonCommercial-ShareAlike 4.0 (CC BY NC SA), la inclusión y difusión del texto completo a través del repositorio institucional de la Universidad Nacional de Colombia y en todas aquellas bases de datos especializadas que el editor considere adecuadas para su indización con miras a incrementar la visibilidad de la revista.
2. Acta Biológica Colombiana permite a los autores archivar, descargar y compartir, la versión final publicada, así como las versiones pre-print y post-print incluyendo un encabezado con la referencia bibliográfica del articulo publicado.
3. Los autores/as podrán adoptar otros acuerdos de licencia no exclusiva de distribución de la versión de la obra publicada (p. ej.: depositarla en un archivo telemático institucional o publicarla en un volumen monográfico) siempre que se indique la publicación inicial en esta revista.
4. Se permite y recomienda a los autores/as difundir su obra a través de Internet (p. ej.: en archivos institucionales, en su página web o en redes sociales cientificas como Academia, Researchgate; Mendelay) lo cual puede producir intercambios interesantes y aumentar las citas de la obra publicada. (Véase El efecto del acceso abierto).