Detection and genome characterization of Potato virus Y isolates infecting potato (Solanum tuberosum L.) in La Union (Antioquia, Colombia)
Detección y caracterización del genoma de aislamientos de Potato virus Y infectando papa (Solanum tuberosum L.) en La Unión (Antioquia, Colombia)
DOI:
https://doi.org/10.15446/agron.colomb.v34n3.59014Keywords:
ELISA, genomics, plant viruses, potyviruses, variants (en)ELISA, genómica, potyvirus, virus de plantas, variantes (es)
Potato virus Y (PVY) is one of the most severe viruses affecting the production of potato (Solanum tuberosum) in the world. This study presents a detailed molecular analysis using nextgeneration sequencing (NGS), IC-RT-qPCR and RT-PCR on the PVY isolates infecting seed-tubers and foliage of potato plants cv. Diacol-Capiro in La Union (Antioquia, Colombia). Analysis of incidence by IC-RT-qPCR in 15 random leaf samples of three cultivation plots and fifteen sprouting tuber eye-buds reveal infection levels between 13.4 and 80%; a higher incidence of 86.7% was observed in seed-tuber samples with threshold cycle (Ct) values as low as 24.3. Genome assembly from a bulk of foliage samples resulted in a consensus PVY genome (PVY_LaUnionF) of 9,702 nt and 399 polymorphic sites within the polyprotein ORF; while the assembled genome from sprouts of tubers has 9,704 nt (PVY_LaUnionT) and contained only six polymorphic nucleotide sites. Phylogenetic analysis demonstrates that the PVY isolates from leaf samples are in the recombinant PVYNTN group (sequence identity >99%); while those from tuber sprouts are in the PVYN/NTN group with identities above 95%. Sanger sequencing of viral capsid suggests the presence of a third variant related to PVYO, a prevalent strain reported in potato fields worldwide.
Recibido: 8 de septiembre de 2016; Aceptado: 30 de noviembre de 2016
ABSTRACT
Potato virus Y(PVY) is one of the most severe viruses affecting the production of potato (Solanum tuberosum) in the world. This study presents a detailed molecular analysis using next-generation sequencing (NGS), IC-RT-qPCR and RT-PCR on the PVY isolates infecting seed-tubers and foliage of potato plants cv. Diacol-Capiro in La Union (Antioquia, Colombia). Analysis of incidence by IC-RT-qPCR in 15 random leaf samples of three cultivation plots and fifteen sprouting tuber eye-buds reveal in fection levels between 13.4 and 80%; a higher incidence of 86.7% was observed in seed-tuber samples with threshold cycle (Ct) values as low as 24.3. Genome assembly from a bulk of foliage samples resulted in a consensus PVY genome (PVY_LaUnionF) of 9,702 nt and 399 polymorphic sites within the polyprotein ORF; while the assembled genome from sprouts of tubers has 9,704 nt (PVY_LaUnionT) and contained only six polymorphic nucleotide sites. Phylogenetic analysis demonstrates that the PVY isolates from leaf samples are in the recombinant PVYNTN group (sequence identity >99%); while those from tuber sprouts are in the PVYN/NTN group with identities above 95%. Sanger sequencing of viral capsid suggests the presence of a third variant related to PVYO, a prevalent strain reported in potato fields worldwide.
Key words:
ELISA, genomics, plant viruses, potyviruses, variants..RESUMEN
El Potato virus Y (PVY) es uno de los virus más severos que afectan la producción de papa (Solanum tuberosum) en el mundo. Este trabajo presenta un análisis molecular detallado utilizando secuenciación de nueva generación (NGS), IC-RT-qPCR y RT-PCR, de aislamientos de PVY procedentes de tubérculo-semilla y tejido foliar de plantas de papa cv. Diacol-Capiro en el municipio de La Unión (Antioquia, Colombia). Las evaluaciones de incidencia de PVY utilizando IC-RT-qPCR en 15 muestras aleatorias de tres lotes, indican niveles vari ables de infección de este virus (13,4 a 80%), mientras que su ocurrencia fue muy alta (86,7%) en 15 muestras de brotes de tubérculo-semilla, con valores de Ct (ciclo umbral) tan bajos como 24,3. El ensamblaje del genoma viral obtenido a partir de una mezcla de muestras de tejido foliar, resultó en una secuencia consenso (PVY_LaUnionF) de 9.702 nt y 399 sitios polimórficos dentro del marco de lectura abierto (ORF) que codifica para la poliproteína. El genoma ensamblado a partir de la mezcla de brotes de tubérculo-semilla presentó un tamaño de 9.704 nt (PVY_LaUnionT) y tan sólo seis sitios polimórficos. Los análisis filogenéticos indican que los aislamientos procedentes de tejido foliar hacen parte del grupo recombinante de PVYNTN (porcentaje de identidad >99%), mientras que los aislamientos de brotes de tubérculos se ubicaron en el clado PVYN/NTN con porcentajes de identidad superiores al 95%. La secuenciación de Sanger de la cápside viral detectó una tercera variante re lacionada con PVYO, una raza con altos niveles de prevalencia en cultivos de papa del mundo.
Palabras clave:
ELISA, genómica, potyvirus, virus de plantas, variantes..Introduction
Potato virus Y (PVY), the type species of the genus Po-tyvirus (Potyviridae), is one the most important viruses affecting the production of potato (Solanum tuberosum L.) worldwide (Karasev and Gray, 2013). Foliar symptoms as sociated to PVY can result in yield reductions between 40% and 70%, while tuber damage significantly compromises its quality and marketability (Nolte et al., 2004; Fageria et al., 2013). PVY can be transmitted mechanically, in a non-persistent manner by aphids such as Myzus persicae or vertically, by infected seed-tubers (Karasev and Gray, 2013). Symptoms induced by PVY are highly dependent on the type of strain and host cultivar and may include leaf mosaics, mottle and crinkling, vein necrosis and necrotic spots; some variants can also induce necrotic ringspots in the tuber skin (Quenouille et al, 2013). Additionally, mixed infection by PVY and Potato virus X (PVX) in several solanaceous crops cause a synergistic interaction that can reduce yields up to 80% (Vance, 1991). PVY can also affect other Solanaceous crops such as tobacco (Nicotiana taba- cum), pepper (Capsicum annumm) and tomato (Solanum lycopersicum) (Scholthof et al., 2011).
PVY virions consist of flexuous particles of about 730 nm in length and a diameter of approximately 11 nm and are composed of a positive-sense single stranded RNA of ap proximately 9,000 nt encapsidated by roughly 2,000 coat proteins subunits (CP) (Ivanov et al., 2014); a covalently linked virus-encoded protein (VPg) is bound to the 5' end (Dougherty and Carrington, 1998; Revers and Garcia, 2015). The PVY genome encodes for a polyprotein of 3060 3063 amino acids, which after processing by three viral encoded proteases, gives rise ten mature protein products: P1, HC-Pro, P3, 6K1, CI, 6K2, NIa-VPg, NIa-Pro, NIb and CP (Dougherty and Carrington,1998; Revers and Garcia, 2015). Recent studies have shown that potyvirus contain two additional ORFs named PIPO (Pretty Interesting Po-tyviridae ORF) and PISPO (Pretty Interesting Sweet Potato Potyvirus ORF) expressed as a result of RNA polymerase transcriptional slippage (Olspert et al., 2015; Untiveros et al., 2016).
The PVY group consists of at least five strains (PVYO, PVYN, PVYC, PVYZ and PVYE) differentiated by the symp toms induced on specific hosts, serological typing and/or nucleotide sequence (Singh et al., 2008). PVYC and PVYO comprise strains that can induce a hypersensitive resistance response (HR) in potato cultivars with the Nc and Ny genes, respectively (Karasev and Gray, 2013). PVY strains that do not induce HR genes and cause systemic necrosis in tobacco belong to the PVYN group, while strains that fail to induce necrosis in tobacco and do not produce the typi cal symptoms of PVY0 and PVYC in their corresponding cultivars are classified as PVYZ, as this strain elicits the Nz gene (Singh et al., 2008; Kerlan et al., 2011). During the past decades genome sequencing has revealed that many PVY strains such as PVYN:0, PVYN-Wi and PVYNTN are the result of recombination between PVY0 and PVYN (Karasev and Gray, 2013). Recombinant strains have received increased attention as they are associated with tuber necrotic ringspot disease in potato (PVYNTN) and tobacco veinal necrosis (PVYN:0, PVYN-Wi) (0gawa et al., 2008).
Due to the emerging importance of PVY, several stud ies have been conducted in the potato producing regions of Colombia that demonstrated the global presence of PVYN and PVYNTN (Gil et al., 2011; Villamil et al., 2014), in addition to two divergent PVY clades named as I-Col y IV-Col (Gallo et al., 2012; Henao et al, 2013). Villamil et al. (2014) obtained the first complete PVY genome in Colombia using next-generation sequencing (NGS) of siRNA from S. tuberosum plants in Cundinamarca; they also found that Potato yellow vein virus (PYVV) and PVY co-infect potato plants with a prevalence of 21% within the Phureja group and 23% within the Andigena group. A recent study using TAS-ELISA and RT-qPCR, revealed widespread PVY infection in S. tuberosum seed-tubers used as planting material in the municipalities of La Union and Yarumal in the province of Antioquia (Medina et al., 2015). Further work confirmed the generalized presence of PVY infecting potato crops in Yarumal and next-generation sequencing (NGS) resulted in the assembly of a complete PVY genome with phylogenetic affinity to necrotic PVYNTN strains (Muñoz et al., 2016). The aim of this study was to perform a detailed molecular analysis on the PVY isolates infecting commercial seed-tubers and potato plants cv. Diacol-Capiro in three plots from La Union (Antioquia) using ELISA, IC-RT-qPCR, RT-PCR and NGS analysis. The presence of two different PVY strain types with phylogenetic affinities to the PVYN and PVYNTN is demonstrated and their relationship to previously sequenced genome discussed. Sanger sequencing also suggests the presence of PVY0 in this Colombian region.
Materials and methods
Plant samples and IC-RT-qPCR
Immunocapture (IC) was conducted following the method of Wetzel et al. (1992) using the Triple Antibody Sandwich PVY detection kit from Agdia (Elkhart, IN). Fifteen random leaf samples collected in three (A, B and C) commercial potato cv. Diacol-Capiro fields (for a total of 45 samples), and fifteen sprouting eye-buds (1-2 cm) of seed-tuber samples from storage commercial cellars in La Union (Antioquia) were evaluated; these tubers were not related to plants sampled in the field plots. Positive and negative controls consisted of lyophilized plant tissue purchased from Agdia (Elkhart, IN). For cDNA synthesis, 12.5 µiL of sample released after TAS-ELISA using Tris-HCl 10 mM pH 8.0 and 1 % Triton X 100 were used as templa te. The reaction also included 200 U of Maxima Reverse Transcriptase, 1X RT buffer, 0,5 mM dNTPs, 100 pmol Oligo-dT primers and 20 U of RNAse inhibitor (Thermo Fisher Scientific, Waltham, MA) and was incubated at 65°C for 5 min, followed by 50°C for 30 min and 85°C for 5 min in a T3 thermal cycler (Biometra, Germany). For the qPCR step, the Maxima SYBR Green/ROX qPCR Master Mix (2X) kit (Thermo Fisher Scientific, Waltham, MA) was used in 25 µiL of reaction containing 12.5 µiL of mix, 10 µiL DEPC water, 20-100 ng cDNA and 0.3 µiM of speci fic primers PVY-1 FP (5'CCA ATC GTT GAG AAT GCA AAA C 3') and PVY-1 RP (5ATA TAC GCT TCT GCA ACA TCT GAG A 3'), designed by Singh et al. (2013) for specific amplification of PVY strains using RT-qPCR with Taqman probes. Amplification cycles consisted of 10 min at 95°C to activate the polymerase, followed by 35 cycles at 95°C for 15 s and 53°C for 45 s, using a Rotor-Gene Q-5plex Platform (Qiagen, Hilden, Germany); fluorescence was measured after each amplification cycle. The samples were considered positive if they exhibited fluorescence values higher than the threshold before the 35th cycle (Schena et al., 2004). Primer specificity was verified by High Resolution Melting in the 50 and 99°C range and the identity of five amplicons, including the positive control, was confirmed by Sanger sequencing.
Next-generation Sequencing and Sanger sequencing
High-throughput sequencing of the S. tuberosum cv. Diacol-Capiro transcriptome was performed on a sample from a bulk of leaves with rugose mosaics, a symptom typically associated with PVY infection, and previously tested using TAS-ELISA with the Triple Antibody Sandwich detection kit specific for PVY strains (SRA 20001/0096) from Agdia (Elkhart, IN). Samples were ground in liquid nitrogen and 100 mg of the remaining powder were used for RNA extraction with the GeneJET Plant RNA Purification Mini Kit (Thermo Fisher Scien tific, Waltham, MA). The library was constructed with the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA) and rRNA depleted with the TruSeq Stranded Total RNA with Ribo-Zero Plant kit (Illumina, San Diego, CA). Sequencing was performed with the Illumina HiSeq 2000 system service provided by Macrogen (Seoul, South Korea). A similar procedure was followed for the tuber transcriptome; in this case, a bulk sample consisting of ten asymptomatic sprouting eye-buds (1-2 cm) from different Diacol-Capiro potato seed-tubers were used. Adapter sequences and low quality bases were removed with SeqTK (https://github.com/lh3/seqtk). Sequence assembly was performed with Trinity (Grabherr et al., 2011) and the contigs corresponding to PVY genome were identified by a local BLASTN search. Both PVY genomes assemblies were confirmed by mapping with Bowtie2 (Langmead and Salzberg, 2012) and checked for incon sistencies and assembly artifacts with Tablet (Milne et al., 2010). Consensus sequences were deposited in GenBank under accession codes KR149260 (PVY_LaUnionF) and KX531041 (PVY_LaUnionT). Additionally, the CP (801 nt) region was amplified by RT-PCR in six symptomatic foliage samples obtained from single plants (LU1, LU2, LU3, LU4, LU5 and LU6) from La Union using primers CP PVYCPF (5ACC ATC AAG SAA ATG ACA CA 3') and PVYCPR (5' CGG AGA GAC ACT ACA TCA CA 3') (Glais et al., 2002) following the protocol reported by Henao et al. (2013) and sequenced by the Sanger method (GenBank accession codes KR902698-KR902703) after gel purification using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific, Waltham, MA).
Bioinformatic analysis
0RFs codifying for viral proteins were identified with BLASTX (Gish and States, 1993). The phylogenetic analy sis was performed in MEGA6 (Tamura et al., 2013) by the Neighbor-Joining method with 1,000 bootstrap replicates using the Tamura 3-parameter method (Tamura, 1992). Rate variation among sites was modeled with a gamma distribution (shape parameter = 0.3). Evolutionary analy ses were conducted in MEGA6 (Tamura et al., 2013). The rates of non-synonymous substitutions per non-synonymous site (Ka) and synonymous substitution per synonymous site (Ks) were estimated by the Nei-Gojobori method using a sliding window of 14 codons (Nei and Gojobori, 1986). The recombination analysis was performed by measuring the local nucleotide percent identity along the genome using in-house perl scripts and plotted with a sliding window of 100 nucleotides; sequences PVYN (AY8849983), PVY0 (EF026074) and PVYC (AJ439544) were used as parents. Multidimensional scaling analysis was performed with cmdscale function in R (www.R-project.org) with distances calculated by the Kimura 2-parameter method (Kimura, 1980).
Results
IC-RT-qPCR detected PVY in all plots (Tab. 1). In plot A, five plants out of 15 were found to be infected (33.3%) with Ct values in the 25.9 to 33.2 range and melting tempera tures between 76.3 and 76.5°C. Plot B exhibited the highest incidence between foliage samples (12/15, 80%) with Ct values as low as 26.4 and an average Tm of 76.7±0.4°C in agreement with the melting temperatures measured in plot A. 0nly two out of 15 leaf samples tested positive in plot C (13.3%) with Ct values of 30.1 and 31.4; Tm values were 76.8 and 78.1, respectively. In contrast, the incidence of PVY in tuber samples was 80% (out of 15 samples) with Ct values as low as 23.7 and an average melting temperature of 77.3±0.9°C. An overview of the Tm data reveals a 1.8°C difference between the extreme values (76.3°C and 78.1°C), suggesting the presence of sequence variants.
1Ct: threshold cycles below 35 were considered positive. 2Tm: melting temperatures.TABLE 1: IC-RT-qPCR detection of PVY in leaf samples from three potato cv. Diacol-Capiro cultivating plots (A-C) and sprouting eye-buds of tubers (T) from La Union (Antioquia).
Plot A
Plot B
Plot C
Tubers
CT1
Tm
2
(°C)
Ct
Tm (°C)
Ct
Tm (°C)
Ct
Tm (°C)
A1
>35
-
B1
>35
-
C1
>35
-
T1
28.1
77.8
A2
>35
-
B2
29.8
77.2
C2
>35
-
T2
27.1
77.2
A3
>35
-
B3
26.4
76.4
C3
>35
-
T3
30.9
76.6
A4
30.3
76.5
B4
29.1
77.1
C4
>35
-
T4
28.2
78.1
A5
>35
-
B5
29.2
77.2
C5
>35
-
T5
27.4
77.4
A6
28.7
76.5
B6
30.4
77.1
C6
>35
-
T6
27.2
77.6
A7
33.2
76.3
B7
>35
-
C7
>35
-
T7
30.1
77.5
A8
>35
-
B8
28.8
76.4
C8
>35
-
T8
>35
-
A9
>35
-
B9
28.7
76.3
C9
>35
-
T9
32.1
78.1
A10
>35
-
B10
28.3
76.9
C10
30.1
78.1
T10
31.1
77.5
A11
28.2
76.5
B11
29.6
76.3
C11
>35
-
T11
>35
-
A12
>35
-
B12
28.2
76.5
C12
31.4
76.8
T12
>35
-
A13
>35
-
B13
>35
-
C13
>35
-
T13
24.3
77.4
A14
25.9
76.3
B14
29.4
76.56
C14
>35
-
T14
23.7
78.1
A15
>35
-
B15
29.6
76.86
C15
>35
-
T15
25.7
77.6
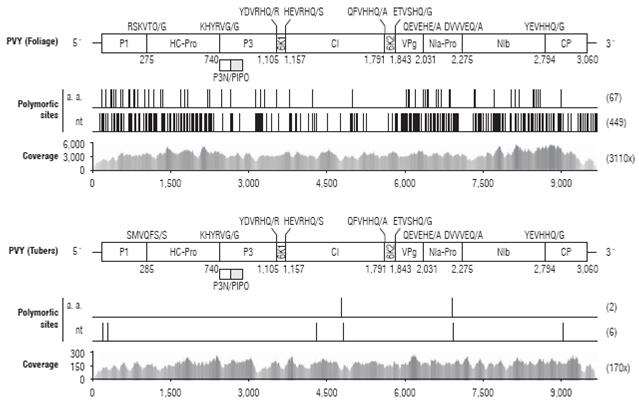
To get a more detailed characterization of the PVY strains infecting S. tuberosum cv. Diacol-Capiro in La Union, a high-throughput transcriptome sequencing was carried out on bulks from both foliage and sprouting eye-buds from tuber samples; PVY infection was confirmed by ELISA prior sequencing. A total of4,754,725 paired-end reads were obtained from the infected leaf-sample with 7,750 reads per million (RPM) attributed to PVY. Assembly resulted in a consensus PVY genome (PVY_LaUnionF) of 9,702 nt (excluding the poly-A tail) with an average sequence depth of 3,110x (Fig. 1A). The consensus sequence contains 449 polymorphic sites with a transition/transversion ratio of 0.85; the high proportion of polymorphisms suggests the presence of mixed variants within the sample, which is not a surprinsing result when bulk samples are used for sequencing. The 0RF encoding the polyprotein is located at positions 188 and 9,373; frameshift product P3N-PIP0 is predicted at nucleotide positions 2,474-2,926. The poly protein 0RF contains 399 polymorphic sites that translate into 67 amino acid changes; as expected, most substitutions map to the third codon position (316). Percentagewise, most changes are amino acid changes are located in the 6K1 (7/52, 13.5%), 6K2 protein (5/52, 9.6%), P1 (25/285, 8.8%) and P3 (25/364, 6.8%). CP (3/267, 1.1°%) and NIb (13/519, 2.5%) showed the lowest degrees of variation. Isolate PVY_ LaUnionF shares 99.8% sequence identity with a similar isolate (PVY_Yarumal: KT336551) from the municipality of Yarumal in the north of Antioquia with phylogenetic affinity to PVYNTN strains (Muñoz et al., 2016).
FIGURE 1: Sequence coverage and mapping of the PVY genomes infecting potato foliage and sprouting eye-buds of tubers in La Union (Antioquia). The relative position of each mature protein in PVY is show on top of each panel; viral protease cleavage sites are shown. The PVY isolated from bulked foliage (PVY_LaUnionF, KR149260) exhibits a very high sequence coverage (3110 x) and variability with 449 polymorphic sites suggesting the presence of mixed variants in the bulked sample. In contrast, the PVY strain from a bulk of tuber sprouts (PVYLaUnionT, KX531041), resulted from an assembly with 170x coverage and only six polymorphisms.
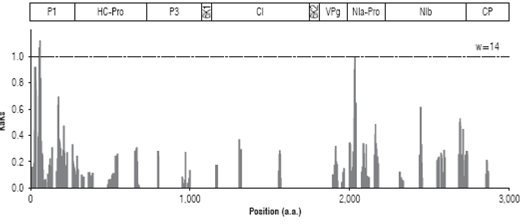
The high levels of variation observed in the PVY foliage assembly may be indicative of sequences experiencing di versifying selection. To address this point, the ratio (Ka/Ks) of non-synonymous substitutions per non-synonymous site (Ka) to synonymous substitution per synonymous site (Ks) was measured along the polyprotein coding region using a sliding window of 14 codons (Fig. 2). Ka/Ks values could be determined for 1,831 sites; 1,473 (80.4%) were under strong purifying selection (Ka/Ks=0), 355 were under weak puri fying selection (19.4%, 0< Ka/Ks <1) and three sites under either neutral or weak positive selection (Ka/Ks >1). All windows under positive selection were located within the P1 protein and centered at amino acid positions 59 (Ka/ Ks=1.05), 60 (Ka/Ks=1.0) and 63 (Ka/Ks=1.09), comprising the amino acid sequence (I/V)RTSK(N/S)G(T/A). A region centered at residue 2,083 within NIa-Pro showed a Ka/Ks very close to the threshold value (0.97) and comprises a region with the sequence (H/D)LFKSLNGS(T/M)E(A/V).
FIGURE 2: Sliding window analysis of genome variants of the PVY consensus isolated from potato foliage in La Union (Antioquia). A window size of 14 was used. Ka/Ks ratios larger than one are indicative of positive selection.
The tuber transcriptome, on the other hand, resulted in a data set comprising 4,754,725 paired-end reads of which 8,256 (1,736 RPM) were attributed to PVY. The assembled genome consists of a 9,704 nt contig (excluding the polyA tail) with average sequence depth of 170x and was named PVY_LaUnionT (Fig. 1B). In contrast to the leaf isolate, the tuber PVY consensus contained only six polymor phic nucleotide sites; two of them, U4746G and T6875C, result in amino acid changes in CI (V1520G) and NIa-Pro (W2230R). The closest PVY isolate available in GenBank corresponds to isolate mar7 (KR270797) infecting S. lycopersicum at the municipality of El Peñol (Muñoz-Baena et al., 2016). Isolate mar7 has phylogenetic affinity to the PVYN group and shares 99.4% nucleotide sequence identity with PVY_LaUnionT. Pairwise comparison among the ge nomes from foliage and tubers reveal significant differences in nucleotide and amino acid sequence identity between regions encoding each of the mature proteins (Table 2). The highest differences were found at the 6K2 and P3 proteins with nucleotide (amino acid) percent identities of 85.9 (94.0) and 92.2 (92.4); the best conserved segment correspond to CP (95.6/99.2) and NIb (95.1/98.5). 5' and 3' untranslated regions share 87.5% and 86.6% nucleotide sequence identity, respectively.
TABLE 2: Pairwise sequence comparison between PVYLaUnionF (KR149260) and PVYLaUnionT (KX531041). Values correspond to the percent identity between the consensus sequences at the nucleotide (nt) and amino acid level (a.a.).

Phylogenetic analysis of complete PVY sequences con firmed that PVY_LaUnionF and PVY_LaUnionT cluster in two different clades within the N/NTN group (Fig. 3A). PVY_LaUnionF is part of a group comprising (only) PVYNTN recombinant strains and its close evolutionary relationship with the PVY isolate from Yarumal becomes clear as they cluster together within the clade. The tuber strain, on the other hand, belongs to a group comprising PVYN strains and other PVYNTN strains; as expected, isolate mar7 from tomato also belongs to this group. The recombinant nature of the PVY_LaUnionF was further confirmed by recombination analysis (Fig. 3B). Sequence comparison with PVYN, PVY0 and PVYC strains reveal three recombination breakpoints at the HC-Pro/P3 and 6K2/NIa-VPg junctions and near the end of the CP region, a pattern typical of PVYNTN strains (recombinant junction: NTNa) (Karasev and Gray, 2013). Recombination analysis of the PVY tuber strain failed to reveal recombination breakpoints (Fig. 3C).
FIGURE 3: Phylogenetic and recombination analysis of the PVY isolates from foliage and tubers in La Union (Antioquia). The phylogenetic tree (A) suggest that the foliage isolate has phylogenetic affinity with a clade composed of recombinant PVYNTN strains while PVY from tubers is more clo sely related to strains of the non-recombinant pvyn/ntn group. Complete PVY genomes from Colombia are shown in bold. Panels B and C show the analysis of recombination for both strains using PVYN, PVYO and PVYC as parents.
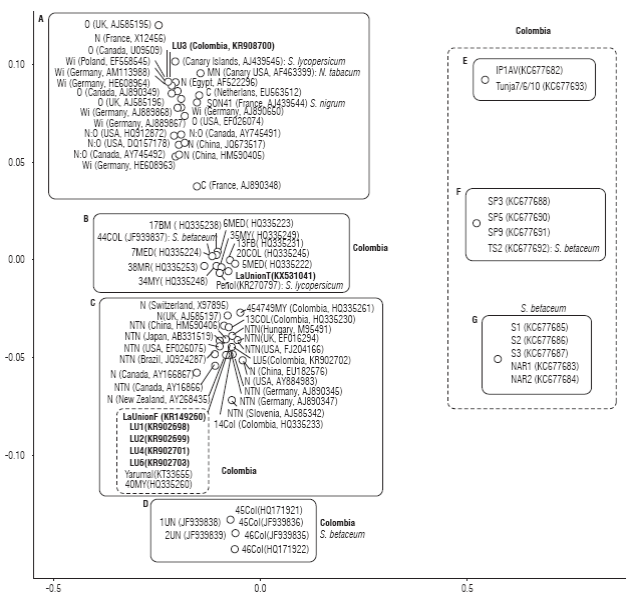
To corroborate the diversity of PVY strains infecting S. tuberosum (cv. Diacol-Capiro) in La Union, partial Sanger sequencing of the CP region was performed in single leaf samples from different commercial cultivation plots (LU1-6). Multidimensional scaling analysis using sequences from Colombia and the rest of the world reveals seven distinct PVY clusters (Fig. 4). Cluster A includes sequences belonging to strains PVYN, PVYN:O, PVYW, PVYC and to NP isolates (non-potato isolates) from all over the world; Cluster C includes sequences corresponding to strains PVYN and PVYNTN; the remaining clusters are composed exclusively from Colombian isolates. The CP segment from PVY_La Union F clustered in group C, which includes the PVY genome from Yarumal, in agreement with phyloge-netic analysis discussed above. This group also includes Sanger sequences LU1, LU2, LU4, LU5 and LU6 and Colombia isolates 13Col, 14Col, 40MY and 454749MY previosuly deposited in GenBank by Gil et al. (2011) and Henao et al. (2013). Group C comprises only PVY strains with S. tuberosum as isolation host. The CP segment of PVY_La Union T clustered in group B composed exclusively by Colombian isolates and includes isolates mar7 (Munoz-Baena et al., 2016) and 44Col (Jaramillo et al., 2011) infect ing tomato and tamarillo (Solanum betaceum) respectively; none of the Sanger sequences obtained here was included within this group. Interestingly, sequence LU3 clustered in group A, which corresponds to the ordinary strain of PVY (PVYO) and suggests the presence of at least a third PVY strain in La Union. No sequences related to groups D-G were found in this study.
FIGURE 4: Multidimensional scaling visualization of partial PVY capsid sequences. Scale in the diagram directly reflects the genetic distances bet ween sequences. When a PVY isolate infects a host different to S. tuberosum the corresponding host is indicated. Sanger sequences obtained in this work are shown in bold.
Discussion
In this work, we have demonstrated the presence of at least two types of PVY strains infecting S. tuberosum (cv. Diacol-Capiro) in the municipality of La Union, the main potato producer of Antioquia. The isolate PVY_LaUnionT, infecting seed-tuber material has phylogenetic affinity with strains of the tobacco veinal necrosis PVY group, PVYN (Singh et al., 2008). PVYN isolates were reported for the first time in Peru and Bolivia in the 1940s (Nobrega and Silber schmidt, 1944) followed by reports in the United States and Europe in the 1960s (Kahn and Monroe, 1963). Symptoms caused by PVYN are not considered to be severe on potato (Singh et al., 2008). Interestingly, Colombian PVYN isolates form a very distinct cluster with widespread distribution in Antioquia and the rest of the country (Gil et al., 2011; Henao et al., 2013; Villamil et al., 2014). Isolates from this group have also been collected in the provinces of Nariño (17BM, 34MY, and 35MY), Boyacá (17BM) and Cundina-marca (13FB). In Antioquia, members of this group have been collected in the municipalities of Santa Rosa de Osos (6MED), La Ceja (38MR) and Sonsón (5MED), the latter two located in the vicinity of La Union. Group C seems to be able to cross the host barrier as attested by isolates 44Col and mar7 infecting S. betaceum and S. lycopersi- cum, respectively (Jaramillo et al., 2011; Muñoz-Baena et al., 2016). PVY_LaUnionT is very closely related to isolate mar7, which together with the geographical proximity of the municipality of El Peñol, suggests that passage of PVY between alternate hosts might be a result of inadequate crop management practices. The infection of PVYNTN and PVYN in tomato has been previously reported in Spain and Poland (Aramburu et al., 2006; Hasiów-Jaroszewska et al., 2015).
The PVY genome from foliage, PVY_La Union F, is clearly a member of the recombinant PVYNTN group of strains and its similarity to a previously sequenced genome from Yarumal in north Antioquia indicate that both isolates probably have a common origin. Analysis of the CP region shows that PVY_La Union F is part of a group of PVY strains with worldwide distribution and previously reported in Colombia in Chipaque (Cundinamarca) (Villamil et al., 2014) and the east of Antioquia in the municipalities of Carmen de Viboral (454749MY), Sonsón (13Col and 14Col) and La Ceja (40MY) (Gil et al., 2011; Henao et al., 2013). The majority of Sanger sequences (LU1-2 and LU4-6) clustered within this group thus confirming the high prevalence of this strain in the potato cultivating regions of eastern Antioquia. Our data indicates that isolate PVY_LaUnionF has high sequence variability and comprises a larger set of sequence variants that PVY_Yarumal, in which only fifteen nucleotide polymorphic sites were observed that translate into three of amino acid changes in P1 (K30E), 6K1 (M1121I) and NIb (K2758R) (Muñoz et al., 2016). The K30E was also found in the PVY_LaUnionF and a M1121V change was observed instead of M1121I; no polymorphism equivalent to K2758R was detected in PVY_LaUnionF. Unfortunately, the genome sequence reported by Villamil et al. (2014) is not available in GenBank and could not be compared with the sequences reported in this work.
The results obtained from the sliding window compari son of non-synonymous to synonymous substitution is in agreement with previous reports showing that P1, P3, 6K1, 6K2, and CP genes have more neutral codons than expected (Cuevas et al., 2012). According to our data, P1 is undergo ing moderate positive selection in the segment comprising residues 59 to 66. A recent population analysis of PVY from Iran identified that 22 sites in P1 and three in NIa-Pro underwent positive selection (Pourrahim and Farzadfar, 2016). Protein P1 is a serine protease involved in RNA silencing and it has been suggested that modulation of P1 activity has evolved to keep viral amplification below host-detrimental levels therefore helping to maintain a higher long-term replicative capacity (Pasin et al., 2014). It would be interesting if future studies could determine whether slight positive selection observed at the N-terminus of P1 in PVY_LaUnionF is related to fine-tuning of its protease function. The other region undergoing relaxed selection pressure is located at the N-terminus of a second protease, i.e., NIa-Pro, which is required for the proteolytic process ing of the most potyviral proteins and has an important effect on infectivity (Ivanov et al., 2014).
Interestingly, sequence analysis of the CP region of sample LU3 reveals that this isolate is most similar to isolates CO1960 (99.7 %), CO1827 (99.7%), CO1801 (99.5%) from the United States (Karasev et al., 2011). These isolates are part of a PVYO group of strains reported to be spreading across the United States and other countries world-wide. The PVYO group can cause mild mosaic in tobacco and induces necrosis and severe stunting in potato cultivars carrying the Ny gene, such as Desiree and Maris Bard, as a result of eliciting the hypersensitive response (HR) (Karasev et al., 2011). This is the first report in Colombia for this group.
In 2014, Colombia had an estimated yield of 19.9 t ha-1 which falls short to the productivity seen in countries such as UK (33.17 t ha-1) and the United States (51 t ha-1) (http://faostaT3.fao.org). It is likely that these low yields are related to the high incidence of viral infections, which might be caused by the lack of adequate seed-certification programs and malpractices in the handling of crops. One of the principal means of virus distribution is through move ment of infected propagation material and once a pathogen gets introduced into a new region it may spread rapidly through the action of vectors and cultural malpractices. PVY is a constant threat to potato production as a result of the lack of efficient resistance of local varieties to many of its strains and, for this reason the main control strategy is based on the certification of tuber seed production. The high incidence levels of PVY detected in foliage and seed-tuber samples and the finding of at least three different strains in a single potato-producing municipality in Antioquia reveal a worrying situation regarding the sanitary conditions of this crop. Urgent measures are required to strengthen current seed-certification programs such as the incorporation of detection methods as IC-RT-qPCR that offer higher sensitivity than ELISA and a constant moni toring of the genomic characteristics of the PVY strains present in each potato-producing region. The latter is of the utmost importance as genome information can help to predict the epidemiological impact of PVY infection, since it is possible to associate the strain genotypes with their impact on yield and tuber quality as well as with alternate hosts and transmission efficiency by arthropod vectors.
Conclusions
The genomes of two different PVY infecting S. tuberosum in the municipality of La Union were obtained using NGS of leaves and tuber sprouts. These genomes comprised 9,072 and 9,704 nt, respectively and encoded a polyprotein of 3060 residues.
The consensus genome from bulked samples of the infected leaves was highly polymorphic containing 399 variable sites at the nucleotide level which leads to 67 amino acid changes in polyprotein, suggesting the presence of mixed variants within the sample. The genome from infected tubers showed little variability as only six nucleotide sub stitutions and two amino acid changes were observed. These differences in variability might be a consequence of different transmission methods occurring in foliar tis sue (e.g. mechanical transmission, vectors, seed-tuber) in comparison with just the primary inoculum of seed-tuber.
High incidence of PVY were observed by IC-RT-qPCR in fifteen commercial seed-tuber samples from la Union (86.7%) and in two out of three potato cv. Diacol-Capiro producing plots (33.4 and 80%). These results underscore the necessity of comprehensive reforms to currently avail able seed-certification programs in Colombia using more sensitive and specific virus detection methods such as real-time RT-PCR.
Acknowledgments
This work was funded by Universidad Nacional de Colom bia (Grant VRI: 34585) and International Foundation for Science (Sweden, Grant C/4634-2).
Literature cited
References
Aramburu, J., L. Galipienso, and M. Matas. 2006. Characteriza tion of Potato virus Y isolates from tomato crops in northeast Spain. Eur. J. Plant Pathol. 115(2), 247-258. Doi: 10.1007/s10658-006-9003-x
Cuevas, J.M., A. Delaunay, M. Rupar, E. Jacquot, and S.F. Elena. 2012. Molecular evolution and phylogeography of Potato virus Y based on the CP gene. J. Gen. Virol. 93(11), 2496-2501. Doi: 10.1099/vir.0.044347-0
Dougherty, W.G. and J.C. Carrington. 1998. Expression and func tion ofpotyviral gene products. Annu. Rev. Phytopathol. 26(1), 123-143. Doi: 10.1146/annurev.py.26.090188.001011
Fageria, M.S., M. Singh, U. Nanayakkara, Y. Pelletier, X. Nie, and D. Wattie. 2013. Monitoring current-season spread of Potato virus Y in potato fields using ELISA and real-time RT-PCR. Plant Dis. 97(5), 641-644. Doi: 10.1094/PDIS-03-12-0283-RE
Gallo, Y., P. Gutiérrez and M. Marín. 2012. Generación de anticuer pos policlonales para la detección de la variante genotípica GIII de PVY, en cultivos de tomate de árbol y papa de Colombia. Rev. Colomb. Biotecnol. 14(1), 245-255.
Gil, J.F., J.M. Cotes, and M. Marín. 2011. Incidencia de potyvirus y caracterización molecular de PVY en regiones productoras de papa (Solanum tuberosum L.) de Colombia. Rev Col. Biotecnol. 13(1), 85-93.
Gish, W. and D.J. States. 1993. Identification of protein coding regions by database similarity search. Nature Genet. 3(3), 266-272. Doi: 10.1038/ng0393-266
Glais, L., M. Tribodet, and C. Kerlan C. 2002. Genomic variability in Potato potyvirus Y (PVY): evidence that PVYNW and PVYNTN variants are single to multiple recombinants between PVYO and PVYN isolates. Arch. Virol. 147(2), 363-378. Doi: 10.1007/s705-002-8325-0
Grabherr, M.G., B.J. Haas, M. Yassour, J.Z. Levin, D.A. Thompson, I. Amit, X. Adiconis, L. Fan, R. Raychowdhury, Q. Zeng, Z. Chen, E. Mauceli, N. Hacohen, A. Gnirke, N. Rhind, F. di Palma, BW. Birren, C. Nusbaum, K. Lindblad-Toh, N. Friedman, and A. Regev. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29(7), 644-652. Doi: 10.1038/nbt.1883
Hasiów-Jaroszewska, B., J. Stachecka, J. Minicka, M. Sowinski, and N. Borodynko. 2015. Variability of Potato virus Y in tomato crops in Poland and development of a reverse-transcription loop-mediated isothermal amplification method for virus detection. Phytopathol. 105(9), 1270-1276. Doi: 10.1094/PHYTO-08-14-0219-R
Henao E., P. Gutiérrez, and M. Marín. 2013. Análisis filogenètico de aislamientos del Potato virus Y (PVY) obtenidos en cultivos de papa (Solanum tuberosum) y tomate de árbol (Solanum betaceum) en Colombia. Actu. Biol. 35(99), 219-232
Ivanov, K.I., K. Eskelin, A. Löhmus, and K. Mäkinen. 2014. Molecu lar and cellular mechanisms underlying potyvirus infection. J. Gen. Virol. 95(7), 1415-1429. Doi: 10.1099/vir.0.064220-0
Jaramillo, M., P. Gutiérrez, L.E. Lagos, J.M. Cotes, and M. Marín. 2011. Detection of a complex of viruses in tama-rillo (Solanum betaceum) orchards in the Andean region of Colombia. Trop. Plant Pathol. 36(3), 150-159. Doi: 10.1590/S1982-56762011000300003
Kahn, R.P. and R.I. Monroe. 1963. Detection of tobacco veinal necrosis (strain of Potato virus Y) in Solanum carensaii and S. andigenum introduce into the United States. Phytopathol. 53, 1356-1359.
Karasev, A.V., X. Hu, C.J. Brown, C. Kerlan, O.V. Nikolaeva, J.M. Crosslin and S.M. Gray. 2011. Genetic diversity of the ordinary strain of Potato virus Y (PVY) and origin of recombinant PVY strains. Phytopathol. 101(7), 778-785. Doi: 10.1094/PHYTO-10-10-0284
Karasev, A.V. and S.M. Gray. 2013. Continuous and emerging chal lenges of Potato virus Y in potato. Annu. Rev. Phytopathol. 51(1), 571-586. Doi: 10.1146/annurev-phyto-082712-102332
Kerlan, C., O.V. Nikolaeva, X. Hu, T. Meacham, S.M. Gray, and A.V. Karasev. 2011. Identification of the molecular make up of the Potato virus Y strain PVYZ: Genetic typing of PVYZ-NTN. Phytopathol. 101(9), 1052-1060. Doi: 10.1094/PHYTO-11-10-0317
Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16(2), 111-120
Langmead, B. and S. Salzberg. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9(4), 357-359. Doi: 10.1038/nmeth.1923
Medina, H.C., P. Gutiérrez, and M. Marín. 2015. Detección del Potato virus Y (PVY) en tubérculos de papa mediante TAS-ELISA y qRT-PCR en Antioquia (Colombia). Bioagro 27(2), 83-92
Milne, I., M. Bayer, L. Cardle, P. Shaw, G. Stephen, F. Wright, and D. Marshall. 2010. Tablet-next generation sequence assem bly visualization. Bioinform. 26(3), 401-402. Doi: 10.1093/bioinformatics/btp666
Muñoz-Baena, L., P. Gutiérrez, and Marín-Montoya. 2016. Detección y secuenciación del Potato virus Y (PVY) que infecta plantas de tomate en Antioquia (Colombia). Bioagro 28(2), 69-80.
Muñoz, D., P. Gutiérrez, and M. Marín. 2016. Detección y caracteri zación molecular del Potato virus Y (PVY) en cultivos de papa (Solanum tuberosum L.) del norte de Antioquia, Colombia. Rev. Prot. Veg. 31(1), 1-11.
Nei, M. and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3(5), 418-426.
Nobrega, N.R. and K. Silberschmidt. 1944. Sobre una provavel variante do virus " Y " da batatinha que tem a peculiaridade de provocar necroses em plantas de fumo. Arq. Inst. Biol. 1944, 307-330.
Nolte, P., J.L. Whitworth, M.K. Thornton, and C.S. McIntosh. 2004. Effect of seedborne Potato virus Y on performance of Russet Burbank, Russet Norkotah, and Shepody potato. Plant Dis. 88(3), 248-252. Doi: 10.1094/PDIS.2004.88.3.248
Ogawa, T., Y. Tomitaka. Nakagawa, and K. Ohshima. 2008. Genetic structure of a population of Potato virus Y inducing potato tuber necrotic ringspot disease in Japan; comparison with North American and European populations. Virus Res. 131(2), 199-212. Doi: 10.1016/j.virusres.2007.09.010
Olspert, A., B.Y. Chung, J.F. Atkins, J.P. Carr, and A.E. Firth. 2015. Transcriptional slippage in the positive-sense RNA virus fam ily Potyviridae EMBO Reports. 16(8), 995-1004. Doi: 10.15252/embr.201540509
Pasin, F., C. Simón-Mateo, and J.A. García. 2014. The hypervariable amino-terminus of P1 protease modulates potyviral replication and host defense responses. PLoS Pathog. 10(3), e1003985. Doi: 10.1371/journal.ppat.1003985
Pourrahim, R. and S. Farzadfar. 2016. Population analysis of Iranian Potato virus Y Isolates using complete genome sequence. Plant Pathol. J. 32(1), 33-46. Doi: 10.5423/PPJ.OA.07.2015.0144
Quenouille, J., N. Vassilakosa, and B. Moury. 2013. Potato virus Y a major crop pathogen that has provided major insights into the evolution of viral pathogenicity. Mol. Plant Pathol. 14(5), 439-452. Doi: 10.1111/mpp.12024
Revers, F. and J.A. García. 2015. Molecular biology of potyviruses. Adv. Virus Res. 92, 101-199. Doi: 10.1016/bs.aivir.2014.11.006
Schena, L., F. Nigro, A. Ippolito, and D. Gallitelli. 2004. Real-time quantitative PCR: a new technology to detect and study phyto-pathogenic and antagonistic fungi. Eur. J. Plant Pathol. 110(9), 893-908. Doi: 10.1007/s10658-004-4842-9
Scholthof, K.B., S. Adkins, H. Czosnek, P. Palukaitis, E. Jacquot, T. Hohn, B. Hohn, K. Saunders, T. Candresse, P. Ahlquist, C. Hemenway, and G.D. Foster. 2011. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12(9), 938-954. Doi:10.1111/j.1364-3703.2011.00752.x
Singh, R.P., J.P.T. Valkonen, S.M. Gray, N. Boonham, R.A.C. Jones, C. Kerlan, and J. Schubert. 2008. Brief review: The naming of Potato virus Y strains infecting potato. Arch. Virol., 153(1), 1-13. Doi: 10.1007/s00705-007-1059-1
Singh, M., R.P. Singh, M.S. Fageria, X. Nie, R. Coffin, and G. Hawkins. 2013. Optimization of a Real-Time RT-PCR assay and its comparison with ELISA, conventional RT-PCR and the grow-out test for large scale diagnosis of Potato virus Y in dormant potato tubers. Am. J. Potato Res. 90(1), 43-50. Doi: 10.1007/s12230-012-9274-z
Tamura, K. 1992. Estimation of the number of nucleotide substitu tions when there are strong transition-transversion and G + C-content biases. Mol. Biol. Evol. 9(4), 678-687.
Tamura, K., G. Stecher, D. Peterson, A. Filipski, and S. Kumar. 2013. MEGA6: Molecular evolutionary genetics analysis. Mol. Biol. Evol. 30(12), 2725-2729. Doi: 10.1093/molbev/msT197
Untiveros, M., A. Olspert, K. Artola, A.E. Firth, J.F. Kreuze, and J.P.T. Valkonen. 2016. A novel sweet potato potyvirus open reading frame (ORF) is expressed via polymerase slippage and suppresses RNA silencing. Mol. Plant Pathol. 17(7),1111-1123. Doi:10.1111/mpp.12366
Vance, V. 1991. Replication of potato virus X RNA is altered in coinfections with potato virus Y. Virology 182(2), 486-494. Doi:10.1016/0042-6822(91)90589-4
Villamil-Garzón, A., W.J. Cuellar, and M. Guzman-Barney. 2014. Natural co-infection of Solanum tuberosum crops by the Potato yellow vein virus and potyvirus in Colombia. Agron. Colomb. 32(2), 213-223. Doi: 10.15446/agron.colomb.v32n2.43968
Wetzel, T., T. Candresse, G. Macquaire, M. Ravelonandro, and J. Dunez. 1992. A highly sensitive immunocapture polymerase chain reaction method for plum pox potyvirus detection. J. Vi rol. Methods. 39(1-2), 27-37. Doi: 10.1016/0166-0934(92)90122-T
How to Cite
APA
ACM
ACS
ABNT
Chicago
Harvard
IEEE
MLA
Turabian
Vancouver
Download Citation
CrossRef Cited-by
1. Andrea Sierra Mejía, Yuliana Marcela Gallo García, Pablo Andrés Gutiérrez Sánchez, Mauricio Alejandro Marín Montoya. (2019). Diseño de cebadores específicos para la detección por RT-PCR del Potato Virus Y (PVY). Acta Biológica Colombiana, 24(3), p.561. https://doi.org/10.15446/abc.v24n3.76523.
2. Lizzete Romero, Huan Peng, Ru Jiang, Wenkun Huang, Celsa García, Joan Gutierrez, Ricardo Holgado, Deliang Peng. (2025). Cytochrome b Gene of Globodera pallida Populations from Colombia Reveals Diversity Within Populations. Phytopathology®, 115(7), p.869. https://doi.org/10.1094/PHYTO-12-24-0395-R.
3. Melissa Riascos Chica, Pablo Andrés Gutiérrez Sánchez, Mauricio Alejandro Marín Montoya. (2018). Identificación molecular de Potyvirus infectando cultivos de papa en el oriente de Antioquia (Colombia). Acta Biológica Colombiana, 23(1), p.39. https://doi.org/10.15446/abc.v23n1.65683.
4. Andrea Sierra, Yuliana Gallo, Meike Estrada, Pablo Gutiérrez, Mauricio Marín. (2021). Detection of four RNA viruses in commercial and informal potato seed tubers in Antioquia (Colombia). Archives of Phytopathology and Plant Protection, 54(5-6), p.273. https://doi.org/10.1080/03235408.2020.1829424.
5. Erika Corrales-Cabra, Mónica Higuita, Rodrigo Hoyos, Yuliana Gallo, Mauricio Marín, Pablo Gutiérrez. (2021). Prevalence of RNA viruses in seeds, plantlets, and adult plants of cape gooseberry (Physalis peruviana) in Antioquia (Colombia). Physiological and Molecular Plant Pathology, 116, p.101715. https://doi.org/10.1016/j.pmpp.2021.101715.
6. Pablo Gutiérrez, Ary Rivillas, Daniel Tejada, Susana Giraldo, Andrea Restrepo, María Ospina, Susana Cadavid, Yuliana Gallo, Mauricio Marín. (2021). PVDP: A portable open source pipeline for detection of plant viruses in RNAseq data. A case study on potato viruses in Antioquia (Colombia). Physiological and Molecular Plant Pathology, 113, p.101604. https://doi.org/10.1016/j.pmpp.2021.101604.
7. Kelsie J. Green, Cassandra N. Funke, Jeffrey Chojnacky, Robert A. Alvarez-Quinto, Jose B. Ochoa, Diego F. Quito-Avila, Alexander V. Karasev. (2020). Potato Virus Y (PVY) Isolates from Solanum betaceum Represent Three Novel Recombinants Within the PVYN Strain Group and Are Unable to Systemically Spread in Potato. Phytopathology®, 110(9), p.1588. https://doi.org/10.1094/PHYTO-04-20-0111-R.
8. Ángela Niño, Francisco J. del Toro, Francisco Tenllado, Tomás Canto, Liliana Franco-Lara. (2021). Molecular insights on potato yellow vein crinivirus infections in the highlands of Colombia. Journal of General Virology , 102(6) https://doi.org/10.1099/jgv.0.001604.
9. Andrea García, Mónica Higuita, Rodrigo Hoyos, Yuliana Gallo, Mauricio Marín, Pablo Gutiérrez. (2023). Prevalence of RNA viruses in certified, and informal potato seed tubers in the province of Antioquia (Colombia). Archives of Phytopathology and Plant Protection, 56(1), p.29. https://doi.org/10.1080/03235408.2023.2170208.
Dimensions
PlumX
Article abstract page views
Downloads
License
Copyright (c) 2016 Agronomía Colombiana

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
© Centro Editorial de la Facultad de Ciencias Agrarias, Universidad Nacional de Colombia
Reproduction and quotation of material appearing in the journal is authorized provided the following are explicitly indicated: journal name, author(s) name, year, volume, issue and pages of the source. The ideas and observations recorded by the authors are their own and do not necessarily represent the views and policies of the Universidad Nacional de Colombia. Mention of products or commercial firms in the journal does not constitute a recommendation or endorsement on the part of the Universidad Nacional de Colombia; furthermore, the use of such products should comply with the product label recommendations.
The Creative Commons license used by Agronomia Colombiana journal is: Attribution - NonCommercial - ShareAlike (by-nc-sa)

Agronomia Colombiana by Centro Editorial of Facultad de Ciencias Agrarias, Universidad Nacional de Colombia is licensed under a Creative Commons Reconocimiento-NoComercial-CompartirIgual 4.0 Internacional License.
Creado a partir de la obra en http://revistas.unal.edu.co/index.php/agrocol/.