Publicado
MetS: an interactive application to identify metabolites in liquid chromatography-mass spectrometry experiments with data independent acquisition
MetS: una aplicación interactiva para identificar metabolitos en experimentos de cromatografía líquida-espectrometría de masas con adquisición independiente de datos
DOI:
https://doi.org/10.15446/dyna.v92n237.114477Palabras clave:
application, metabolomics, metabolite identification, LC-MS/MS, data-independent acquisition (en)aplicación, metabolómica, identificación de metabolitos, LC-MS/MS, adquisición independiente de datos (es)
Descargas
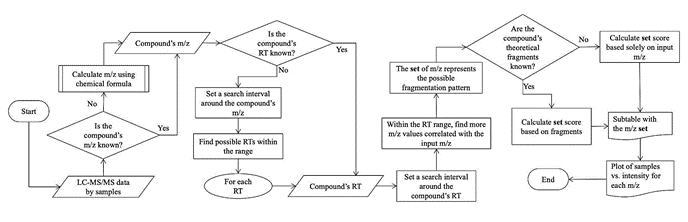
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) with data-independent acquisition (DIA) enables the detection of metabolites in biological samples. However, identifying metabolites from DIA data remains challenging due to the complexity of the data. This work presents "Metabolomic Search" (MetS), a software application developed to facilitate metabolite identification in DIA experiments. The application supports filtering, correlation analysis, and similarity scoring algorithms to match DIA data to user-provided metabolite mass-to-charge ratios and fragmentation patterns. The graphical user interface is straightforward and intuitive, allowing easy data uploading, parameter configuration, and results exploration. Tests on different Solanaceae samples demonstrated successful identification of target metabolites such as scopolamine. By enabling rapid compound screening, MetS can support metabolomic based research in pharmaceutical, biotechnological, and clinical domains. The availability of this open-source tool could help address the pressing need for metabolite annotation in increasingly prevalent DIA experiments.
La cromatografía líquida-espectrometría de masas en tándem (LC-MS/MS) con adquisición independiente de datos (DIA) permite la detección de metabolitos en muestras biológicas. Sin embargo, identificar metabolitos a partir de datos DIA sigue siendo un desafío debido a la complejidad de los datos. Este trabajo presenta "Metabolomic Search" (MetS), una aplicación de software desarrollada para facilitar la identificación de metabolitos en experimentos DIA. La aplicación admite algoritmos de filtrado, análisis de correlación y puntuación de similitud para hacer coincidir los datos de DIA con las relaciones masa-carga de metabolitos y los patrones de fragmentación proporcionados por el usuario. La interfaz gráfica de usuario es sencilla e intuitiva y permite cargar datos, configurar parámetros y explorar resultados fácilmente. Las pruebas en diferentes muestras de Solanaceae demostraron una identificación exitosa de metabolitos objetivo como la escopolamina. Al permitir la detección rápida de compuestos, MetS puede respaldar la investigación basada en metabolómica en los dominios farmacéutico, biotecnológico y clínico. La disponibilidad de esta herramienta de código abierto podría ayudar a abordar la necesidad apremiante de anotación de metabolitos en experimentos DIA cada vez más frecuentes.
Referencias
[1] Yang, Q. et al., Metabolomics biotechnology, applications, and future trends: a systematic review. RSC Adv., 9(64), pp. 37245–37257, 2019. DOI: https://doi.org/10.1039/C9RA06697G
[2] Coskun, O., Separation Tecniques: Chromatography. North Clin Istanbul, 2016. DOI: https://doi.org/10.14744/nci.2016.32757
[3] Amorim-Madeira, P.J., and Helena, M., Applications of tandem mass spectrometry: from structural analysis to fundamental studies. Tandem Mass Spectrometry - Applications and Principles, J. Prasain, Ed., InTech, 2012. DOI: https://doi.org/10.5772/31736
[4] Li, J., Smith, L.S., and Zhu, H.-J., Data-independent acquisition (DIA): an emerging proteomics technology for analysis of drug-metabolizing enzymes and transporters. Drug Discovery Today: Technologies, 39, pp. 49–56, Dec. 2021. https://doi.org/10.1016/j.ddtec.2021.06.006
[5] Perez De Souza L. & Fernie, A. R., Computational methods for processing and interpreting mass spectrometry-based metabolomics. Essays in Biochemistry, 68(1), pp. 5–13, 2024. DOI: https://doi.org/10.1042/EBC20230019
[6] Shen, S., Zhan, C., Yang, C., Fernie, A.R., and Luo, J., Metabolomics-centered mining of plant metabolic diversity and function: past decade and future perspectives. Molecular Plant, 16(1), pp. 43–63, 2023. DOI: https://doi.org/10.1016/j.molp.2022.09.007
[7] Röst H.L. et al., OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol, 32(3), pp. 219–223, 2014. DOI: https://doi.org/10.1038/nbt.2841
[8] Mahieu, N.G., Genenbacher, J.L., and Patti, G.J., A roadmap for the XCMS family of software solutions in metabolomics. Current Opinion in Chemical Biology, 30, pp. 87–93, 2016. DOI: https://doi.org/10.1016/j.cbpa.2015.11.009
[9] Tsugawa, H. et al., MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods, 12(6), pp. 523–526, 2015. DOI: https://doi.org/10.1038/nmeth.3393
[10] Ruttkies, C., Neumann, S., and Posch, S., Improving MetFrag with statistical learning of fragment annotations. BMC Bioinformatics, 20(1), art. 376, 2019. DOI: https://doi.org/10.1186/s12859-019-2954-7
[11] Lai, Z. et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat Methods, 15(1), pp. 53–56, 2018. DOI: https://doi.org/10.1038/nmeth.4512
[12] Cai, Y., Zhou, Z., and Zhu, Z.-J., Advanced analytical and informatic strategies for metabolite annotation in untargeted metabolomics. TrAC Trends in Analytical Chemistry, 158, art. 116903, 2023. DOI: https://doi.org/10.1016/j.trac.2022.116903
[13] Moghe, G., and Strickler, S., metaPathwayMap: a tool to predict metabolic pathway neighborhoods from structural classes of untargeted metabolomics peaks. bioRxiv. Art. 48433, 2022. DOI: https://doi.org/10.1101/2022.03.15.484337
[14] Wang, X. et al. MetMiner: a user‐friendly pipeline for large‐scale plant metabolomics data analysis. JIPB, art. 13774, 2024. DOI: https://doi.org/10.1111/jipb.13774
[15] Abajirov, D., Does functional programming improve software quality?: an empirical analysis of open-source projects on GitHub. Universität Stuttgart. 2021, DOI: https://doi.org/10.18419/OPUS-11582
[16] Chambers, J.M., Object-Oriented Programming, Functional Programming and R. Statist. Sci., 29(2), 2014. DOI: https://doi.org/10.1214/13-STS452
[17] Guo, J., Yu, H., Xing, S., and Huan, T., Addressing big data challenges in mass spectrometry-based metabolomics. Chem. Commun., 58(72), pp. 9979–9990, 2022. DOI: https://doi.org/10.1039/D2CC03598G
[18] Barbier Saint Hilaire, P. et al. Comparative evaluation of data dependent and data independent acquisition workflows implemented on an orbitrap fusion for untargeted metabolomics. Metabolites, 10(4), art. 158, 2020. DOI: https://doi.org/10.3390/metabo10040158
[19] Ledesma-Escobar, C.A., Priego-Capote, F., and Calderón-Santiago, M., MetaboMSDIA: a tool for implementing data-independent acquisition in metabolomic-based mass spectrometry analysis. Analytica Chimica Acta, 1266, art. 341308, 2023. DOI: https://doi.org/10.1016/j.aca.2023.341308
[20] Ebbels, T.M.D. et al. Recent advances in mass spectrometry-based computational metabolomics. Current Opinion in Chemical Biology, 74, art. 102288, 2023. DOI: https://doi.org/10.1016/j.cbpa.2023.102288
[21] Wang, M. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol, 34(8), pp. 828–837, 2016. https://doi.org/10.1038/nbt.3597
[22] Collins, S.L., Koo, I., Peters, J.M., Smith, P.B., and Patterson, A.D., Current challenges and recent developments in mass spectrometry–based metabolomics. Annual Rev. Anal. Chem., 14(1), pp. 467–487, 2021. DOI: https://doi.org/10.1146/annurev-anchem-091620-015205
[23] Ahmad. Software tools in Bioinformatics: a survey on the importance and Issues faced in implementation. Global Engineers and Technologists Review, 3, pp. 6–10, 2013
[24] Mangul, S. et al. Challenges and recommendations to improve the installability and archival stability of omics computational tools. PLoS Biol, 17(6), art. e3000333, 2019. DOI: https://doi.org/10.1371/journal.pbio.3000333
Cómo citar
IEEE
ACM
ACS
APA
ABNT
Chicago
Harvard
MLA
Turabian
Vancouver
Descargar cita
Licencia
Derechos de autor 2025 DYNA

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
El autor o autores de un artículo aceptado para publicación en cualquiera de las revistas editadas por la facultad de Minas cederán la totalidad de los derechos patrimoniales a la Universidad Nacional de Colombia de manera gratuita, dentro de los cuáles se incluyen: el derecho a editar, publicar, reproducir y distribuir tanto en medios impresos como digitales, además de incluir en artículo en índices internacionales y/o bases de datos, de igual manera, se faculta a la editorial para utilizar las imágenes, tablas y/o cualquier material gráfico presentado en el artículo para el diseño de carátulas o posters de la misma revista.
















