Algoritmo para la detección de glóbulos rojos superpuestos en imágenes microscópicas de extendidos de sangre periférica
Algorithm for detection of overlapped red blood cells in microscopic images of blood smears
DOI:
https://doi.org/10.15446/dyna.v83n198.47177Palabras clave:
Procesamiento digital de imágenes, hematología, hough, k-means, superposición, glóbulos rojos, watershed (es)digital image processing, hematology, Hough, k-means, overlap, red blood cells, watershed (en)
Descargas
DOI: https://doi.org/10.15446/dyna.v83n198.47177
Algorithm for detection of overlapped red blood cells in microscopic images of blood smears
Algoritmo para la detección de glóbulos rojos superpuestos en imágenes microscópicas de extendidos de sangre periférica
Miguel Fabián Romero-Rondón a, Laura Melissa Sanabria-Rosas a, Lola Xiomara Bautista-Rozo a & Alfonso Mendoza-Castellanos b
a Escuela de Ingeniería de Sistemas e Informática, Universidad
Industrial de Santander, Colombia. miguelfromeror@gmail.com, melissasanabriarosas@gmail.com, lxbautis@uis.edu.co
b Biosys Ltda, Colombia. almecas99@yahoo.com
Received: November 11th, 2014. Received in revised form: August 20th, 2015. Accepted: March 30th, 2016.
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Abstract
The hemogram
is one of the most requested medical tests as it presents details about the
three cell series in the blood: red series, white series and platelet series.
To make some diagnostics, the specialist must undertake the test manually,
observing the blood cells under the microscope, which implies a great physical
effort. In order to facilitate this work, different digital image processing techniques
to detect and classify red blood cells have been proposed. However, a common
problem is the presence of overlapped cells, which generate various flaws in
the analysis. Therefore, the implementation of an algorithm to address the
problem of red blood cells overlapped in cellular smear images is proposed in
order to support the clinician in the visual reading process. The method was
tested with 50 images in which the indices of sensitivity and specificity were
calculated, and the effectiveness of the algorithm developed was shown.
Keywords: digital image processing; hematology; Hough; k-means; overlap; red blood cells; watershed.
Resumen
El hemograma
es uno de los exámenes médicos más solicitados, ya que ofrece información
detallada sobre las tres líneas celulares presentes en la sangre: la serie
roja, la blanca y la plaquetaria. Para emitir algunos diagnósticos el
especialista debe hacerlo de forma manual, observando en el microscopio las
células sanguíneas, lo que implica mayor esfuerzo. Con el propósito de
facilitar este trabajo, se han propuesto diferentes técnicas de procesamiento
digital de imágenes para la detección y clasificación de glóbulos rojos, pero
se ha encontrado un problema muy común que es la presencia de células
superpuestas, lo cual genera diversos errores en los resultados. Por esta
razón, se propone la implementación de un algoritmo que permita abordar el problema
de superposición de glóbulos rojos en imágenes de frotis celular, con el fin de
dar soporte al especialista en el proceso de lectura visual. El método fue
probado con 50 imágenes, con las cuales se calcularon los índices de
especificidad y sensibilidad, mostrando la efectividad del algoritmo
desarrollado.
Palabras clave: Procesamiento digital de imágenes; hematología; hough; k-means; superposición; glóbulos rojos; watershed.
1. Introduction
Blood is composed of a liquid part known as plasma and formed elements such as red blood cells (RBC), white blood cells (WBC) and platelets [1]. In order for it to be analyzed, a study called a hemogram is undertaken, which counts the number of figurative elements in a certain volume of blood. It also allows specialists to confirm or assume various diseases according to the alterations presented in the count [2].
Currently, this test is performed automatically using specialized equipment and calibrated according to the ranges specified by the manufacturer [3,4]. When the counting is outside this range, the specialists analyze the state of maturation, staining characteristics, the content of granules, inclusions and cellular forms. The observation of cells from a blood smear using a microscope then becomes necessary, which is a tedious process for the individual [5].
In order to facilitate this work and make it more efficient a wide range of algorithms in digital image processing have been developed to capture, represent, analyze and classify objects in microscopic images. Moreover, in automation tasks such as segmentation or classification of elements in an image, it has been found that a common problem is the presence of overlapped cells [6].
Different methods to address the problem of overlapped cells have been proposed in several studies. In [7-12], watershed transformation is used in which over-segmentation problems occur; however, better results can be obtained if sign marking is employed. Therefore, most researchers recommend performing suitable image preprocessing or pre-selection of objects in order to appropriately choose the benchmarks.
In other studies [13-17], separation was performed using techniques based on the objects edge. These take into account its concavity or convexity in order to find the points of maximum curvature and then join them by using a triangulation technique. The errors in these methods can be observed when the cells have an irregular edge.
One way to do the groups separation is to use the morphological operation of erosion, as shown in [18-21]; however, this method is only effective when the convexity of the groups is deep enough. Some alternatives to solve this problem is to erode certain parts of the cell, but this is very limited due to the sensitivity of the parameters, and the computational cost is also high.
Another alternative is proposed by [22-26], who use the Hough transform to detect circular shapes and thus segment images of RBC in peripheral blood smears. This takes advantage of circular shape of erythrocytes.
This article describes the design of an algorithm that uses morphological operations, the watershed method, the Hough transform and the clustering method of k-means to detect overlapped RBC. This will, in turn, smooth out the curve of their edge with Bézier curves. Finally, we will present the specificity and sensitivity rates found in test scores from a group of 50 microscopic images of blood cells.
2. Algorithm design
The proposed algorithm consists of three stages, as shown in Fig. 1. The first stage is the pre-processing stage, in which the removal of white blood cells and platelets is undertaken.
The second step is segmentation, in which the separation of groups of cells to subsequently detect the centers of each is made. Finally, there is a post-processing stage, in which the edges of all RBC appearing in the image are plotted.
2.1. Preprocessing
2.1.1. White blood cell removal
In order to highlight the white blood cells, image preprocessing is performed by converting the image from an RGB color model to an HSV model. The S channel is chosen because it has a higher contrast, which means that platelets and white blood cell nuclei can be clearly distinguished, as is shown in Fig. 2 (b). To expedite the process the image is binarized using the Otsu's method [27] in order to create a black and white image containing the nuclei of white blood cells and platelets (see Fig. 2 (c)).
As can be observed, some noise and the WBC cytoplasm still remain on the image. Therefore, the small objects are removed using a morphological opening; the result is shown in Fig. 2 (d). Subsequently, the image is dilated with a disc-shaped structuring element in order to remove part of the WBC cytoplasm. This result can be seen in Fig. 2 (e).
Finally, white blood cell removal on the original image is shown in Fig. 2 (f).
2.1.2. Platelets removal
As seen in the previous section, the extraction of channel S and binarization also make it possible to distinguish platelets. Therefore these steps are repeated to remove them. Subsequently, a subtraction between the binarized image of S channel and binarization of the original image is performed.
Despite this process, some noise and platelets still remain, and as such, objects with a small area are removed by employing a morphological opening operation with a disc-shaped structuring element.
2.2. Segmentation
2.2.1. Group Segmentations
In order to improve the image contrast, a background template is obtained using a morphological opening over the entire image. It is then subtracted from the grayscale image.
If the full image after background removal is binarized (objects take the value 1 and background 0), the black and white image presented in Fig. 3 (b) is obtained. As a result of this binarization, holes can be seen inside the cells due to lighting and the concave shape of RBC. Thus, these dark spaces are filled using a technique that employs morphological dilation to close the RBC centers. The result of this operation can be seen in Fig. 3 (c).
The noise that is still present in Fig. 3 (c) is removed by using a morphological opening with a disc-shaped structuring element. A morphological closing is then made on the image to close the RBC that were opened in the binarization step due to illumination on its surface; the result is presented in Fig. 3 (d).
Considering that the average circular area of a RBC is 35 micrometers, a sketch can be plotted for the distribution curves of the areas for the individual. There will be two overlapped and three or more overlapped RBC, as shown in Fig. 4.
Misclassification can occur at the intersection of probability distribution curves. To overcome these errors, the following measures are ensured:
- In the detection of a single cell or two overlapped cells, the convex hull is used. If it is equal to the cell edge, the connected component is classified as a single cell; otherwise it is classified as a group of two cells.
- In the classification of groups of two or three cells, the Hough transform is used to detect the number of circles present. If the result differs with minimal difference in the number of cells detected according to the area, Hough determines the amount taken to be true. But, if the difference is greater than 1, the number of cells defined by the area is maintained. This procedure is also useful to classify groups of more than three cells.
Based on this idea, we can classify according to the area of the connected components found in the image in order to gain an understanding of the amount of cells that could be in each one. The result of this classification is shown in Fig. 5; individual cells, groups of two cells, and groups of three or more cells are highlighted in the image.
2.2.2. Centers detection
The watershed transformation is an image segmentation tool that is based on mathematical morphology. The image can be considered to be a topographic relief where the gray
level of each pixel corresponds to a physical elevation. When the landscape is immersed in a lake with holes pierced in local minima, catchment basins fill up with water, starting at these local minima. At points where water coming from different basins meets, dams are built. As a result, the landscape is partitioned into regions or basins separated by dams; these are called watershed lines [28].
The main drawback of this method is the over-segmentation due to the presence of many local minima. To decrease the effect of severe over-segmentation, marker-controlled watershed transformations can be used [29].
For this reason, in this step, different approaches are used in order to define the suitable markers in watershed transform.
Using erosion: After classifying the cell groups we need to find out the centroid of each cell that defines the connected component. As a first attempt to discover this center, successive erosions are used. In order to do this, an image such as Fig. 6(a) is created for each connected component that was not classified as an individual [30].
First, erosions are made iteratively until the connected component is separated. Then, in order to look for the line that allows the cells to be separated we had to find the components' center of mass that were in Fig. 6 (b) in order to use them as markers in the watershed method.
Fig. 6 (c) shows the centroids and watershed lines that are drawn on the connected component.
Using Hough: The problem that arises is that, after erosion, some cells are not separated; however, their morphology was taken to be an advantage (typically RBC have a circular shape) using the Hough transform.
The Hough transform is applicable to any function of the form , where
, where  is a vector of coordinates and
is a vector of coordinates and  is a vector of coefficients. In
Cartesian coordinates, the equation for a circle is given by:
is a vector of coefficients. In
Cartesian coordinates, the equation for a circle is given by:

To detect a circle we need to search
parameter triplets  in a three dimensional
parameter space. The edge pixels are used as the input point for the Hough
transform circle detection.
in a three dimensional
parameter space. The edge pixels are used as the input point for the Hough
transform circle detection.
If the edge pixel lies on a circle, the
locus for the parameters of that circle is a right circular cone surface. This
means that each point  produces a cone surface in
parameter space.
produces a cone surface in
parameter space.
If a set of edge pixels in a cell image
are lying on the circle with parameters , the resultant loci of each point will pass the same point
, the resultant loci of each point will pass the same point  in parameter space. Thus, many
right circular cone surfaces will intersect at a common point [23].
in parameter space. Thus, many
right circular cone surfaces will intersect at a common point [23].
Below, Fig. 7 shows the centers of the circles detected by Hough in groups of two and three cells. Subsequently, using these centers as markers, watershed finds the overlapping lines.
Using K-means: After undertaking the steps above, there are still unseparated cells; many of these have an irregular shape, so it is important to use a method that does not depend on the morphology of the object.
The third technique used is k-means, it is the clustering algorithm used to determine the natural spectral groupings present in a data set. This receives as an input the number of clusters to be located in the data. The algorithm then arbitrarily seeds or locates that number of cluster centers in multidimensional measurement space. Each pixel in the image is then assigned to the cluster whose arbitrary mean vector is closest. The procedure continues until there is no significant change in the location of class mean vectors between successive iterations of the algorithm [31].
The set of points that belong to the edge is passed as a parameter to a k-means algorithm, and it also approximates the center of each cell, as shown in Fig. 8 (a). Again, using the result of k-means as markers, watershed detects overlapping lines. This result is presented in Fig. 8 (b).
2.3. Post-Processing
The purpose of this stage is to draw the cell edge in the overlapping area. First, each cell is separated using the images that come from each of the above steps. Their edge is found and then the endpoints of that edge are obtained. In this step, there are two types of cells; the first is shown in Fig. 9 (a), in which there are only two endpoints.
In this case, Bézier is used to soften the missing edge of the cell. The following are used as control points: 50 points taken from the edge, including 25 points behind each endpoint, as shown in Fig. 10 (b); and an arc which is formed using the two endpoints and the cell's center of mass(see Fig. 10 (c)). The result, presented in Fig. 10 (d), is a smooth curve of the cell edge.
The second type is shown in Fig. 9 (b), in which a cell has more than two endpoints on its edge. For these types of cells, Bézier is not used since it presents several problems: the execution time increases, there are fewer control points and the edge of these objects is very irregular. Therefore, the distance of each point on the edge to the center of mass of the cell was measured, and an average radius was selected to draw a circle in order to define the edge of the cell.
3. Results and discussion
A computer with an Intel Core I3 processor was used for the test. The processing time per image was, on average, 25 seconds. 50 images of 1076 x 840 were selected, in which the three types of blood cells (white, red and platelets) are present; although, some of them are blurred and have noisy spots.
Table 1 shows the rates of specificity and sensitivity to detect groups of two and three cells, as these are the most common in the observation area of peripheral blood smears. The numbers were estimated by comparing with manual counting results.
The sensitivity of the algorithm
quantifies its ability to correctly identify the groups of two and three cells.
In other words, it is the proportion of the true positives  that are correctly identified
by the algorithm, which is given by
that are correctly identified
by the algorithm, which is given by

where 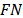 represents the false
negatives, the groups that were not detected. Moreover, the specificity is the
algorithm's ability to not identify as groups of two or three cells the groups
that do not meet these characteristics. In other words, the proportion of the
true negatives
represents the false
negatives, the groups that were not detected. Moreover, the specificity is the
algorithm's ability to not identify as groups of two or three cells the groups
that do not meet these characteristics. In other words, the proportion of the
true negatives  is given by
is given by , where
, where  represents the false
positives: the groups that did not have two or three cells and were detected as
such [32].
represents the false
positives: the groups that did not have two or three cells and were detected as
such [32].

The final output looks like the one in Fig. 11, which shows that the algorithm also manages to classify groups of more than three cells. In addition, Fig. 12 shows comparative results for three different algorithms: a watershed approach, an algorithm using the Hough transform, and our method. The Hough-based algorithm was superior to the watershed approach, but it still could not identify cells with no circular shape. In contrast, our algorithm performs better in cells with an irregular shape, and does not present over-segmentation as the watershed-based algorithm does.
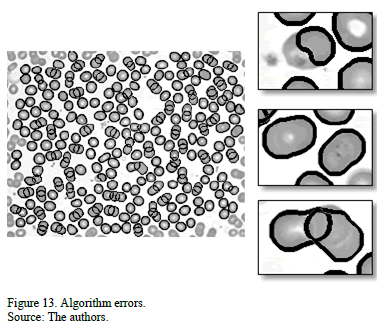
However, Fig. 13 shows some errors in our algorithm. When there is a platelet over the RBC, the preprocessing step deforms the cell, and when the size of the cells is out of the average range, the area classification of the groups is flawed.
Changes in illumination and contrast also significantly affect the results. The light intensity of the microscope Olympus CX-31 was set at 5.7 volts, and the aperture iris diaphragm was set to 80% of the aperture of the objective. For these specific values, in the thresholding step of the nuclei of white blood cells, the value obtained by the Otsu's method increased by 1.5. If the values of illumination and contrast vary, the scaling value should be changed accordingly in order to prevent the result being affected.
In the Red Blood Cell Segmentation step, as the Wright's stain was used, the Green channel of the RGB input image is thresholded as the variations of tone are best appreciated in this channel. If the staining method changes, the segmentation step can give different results.
4. Conclusions
- The illumination of the microscope, the staining of the plate, and properly performing the extent are very important in achieving a correct segmentation.
- It is important that the images are taken in the observation area that is used by the specialist. This is because in this area the shape of RBC allows Hough transform to offer better results since the cells in other regions of the plate are usually deformed.
- Using Watershed, Hough and K-means separately does not equate to employing an effective technique because each one of them has its own limitations. Therefore, an algorithm that takes advantage of each of these techniques to get better results was developed.
- The specificity and sensitivity indices obtained show that the algorithm developed for detection and classification of overlapped red blood cells was highly accurate.
Acknowledgments
The authors express their gratitude to the following entities: Universidad Industrial de Santander (UIS), Grupo de Investigación en Ingeniería Biomédica (GIIB), and Biosys Ltda. for the support offered in undertaking this project.
References
[1] Rogers, K., Blood: Physiology and Circulation. New York: Britannica Educational, 2010
[2] Bijlani, R., Fundamentals physiology a textbook for nursing students. New Delhi: Jaypee Brothers Publishers, 2001.
[3] Estridge, B., Basic Medical Laboratory Techniques. Clifton Park: Cengage Learning, 2000.
[4] Rodak, B., Hematology: Clinical principles and applications. Missouri: Elsevier Health Sciences, 2007.
[5] Ruiz, G., Fundamentos de hematología. Buenos Aires: Ed. Médica Panamericana, 1994.
[6] Kothari, S., Chaudry, Q. and Wang, M., Automated cell counting and cluster segmentation using concavity detection and ellipse fitting techniques, Biomedical Imaging: From Nano to Macro, 2009. pp. 795-798. DOI: 10.1109/ISBI.2009.5193169
[7] Cloppet, F. and Boucher, A., Segmentation of complex nucleus configurations in biological images. Pattern Recognition Letters, 31(8), pp. 755-761, 2010. DOI: 10.1016/j.patrec.2010.01.022
[8] Sharif, J., et al., Red blood cell segmentation using masking and watershed algorithm: A preliminary study, Biomedical Engineering (ICoBE), 2012. pp. 258-262. DOI: 10.1109/ICoBE.2012.6179016
[9] Veta, M., et al., Marker-controlled watershed segmentation of nuclei in H&E stained breast cancer biopsy images, Biomedical Imaging: From Nano to Macro, 2011. pp. 618-621. DOI: 10.1109/ISBI.2011.5872483
[10] Xu, S., Liu, H. and Song, E., Marker-controlled watershed for lesion segmentation in mammograms. Journal of Digital Imaging, 24(5), pp. 754-763, 2011. DOI: 10.1007/s10278-011-9365-2
[11] Di Cataldo, S. et al.. Automated segmentation of tissue images for computerized IHC analysis. Computer Methods and Programs in Biomedicine, 100(1), pp. 1-15, 2010. DOI: 10.1016/j.cmpb.2010.02.002
[12] Karvelis, P., Likas, A. and Fotiadis, D., Identifying touching and overlapping chromosomes using the watershed transform and gradient paths. Pattern Recognition Letters, 31(16), pp. 2474-2488, 2010. DOI: 10.1016/j.patrec.2010.08.002
[13] Wen, Q., Chang, H. and Parvin, B., A Delaunay triangulation approach for segmenting clumps of nuclei, Biomedical Imaging: From Nano to Macro, 2009. pp. 9-12. DOI: 10.1109/ISBI.2009.5192970
[14] Kumar, S. et al., A rule-based approach for robust clump splitting. Pattern Recognition, 39(6), pp. 1088-1098, 2006. DOI: 10.1016/j.patcog.2005.11.014
[15] Wang, H., Zhang, H. and Ray, N., Clump splitting via bottleneck detection and shape classification. Pattern Recognition, 45(7), pp. 2780-2787, 2012. DOI: 10.1016/j.patcog.2011.12.020
[16] Berge, H. et al., Improved red blood cell counting in thin blood smears, Biomedical Imaging: From Nano to Macro, 2011. pp. 204-207. DOI: 10.1109/ISBI.2011.5872388
[17] Latorre, A. et al., Segmentation of neuronal nuclei based on clump splitting and a two-step binarization of images. Expert Systems with Applications, 40(16), pp. 6521-6530, 2013. DOI: 10.1016/j.eswa.2013.06.010
[18] Di Ruberto, C., Dempster, A., Khan, S. and Jarra, B., Analysis of infected blood cell images using morphological operators. Image and Vision Computing, 20, pp. 133-146, 2002. DOI: 10.1016/S0262-8856(01)00092-0
[19] Buggenthin, F. et al., An automatic method for robust and fast cell detection in bright field images from high-throughput microscopy. BMC Bioinformatics, 14(297), pp. 1-12, 2013. DOI: 10.1186/1471-2105-14-297
[20] Prasad, K. et al., Image analysis approach for development of a decision support system for detection of malaria parasites in thin blood smear images. Journal of digital imaging, 25, pp. 542-549, 2012. DOI: 10.1007/s10278-011-9442-6
[21] Amit-Kunar, P., Tembhare, P. and Pote, C., Enhanced identification of malarial infected objects using otsu algorithm from thin smear digital images. International Journal of Latest Research in Science and Technology. 1, pp. 159-163, 2012.
[22] Priyankara, G.P.M. et al., An extensible computer vision application for blood cell recognition and analysis. Thesis, Department of Computer Science and Engineering, University of Moratuwa, Sri Lanka, 2006.
[23] Guan, P.P. and Hong, Y., Blood cell image segmentation based on the Hough transform and fuzzy curve tracing. Machine Learning and Cybernetics (ICMLC), 2011. pp. 1696-1701. DOI: 10.1109/ICMLC.2011.6016961
[24] Prasad, D.K., Leung, M.K. and Cho, S.Y., Edge curvature and convexity based ellipse detection method. Pattern Recognition, 45(9), pp. 3204-3221, 2012. DOI: 10.1016/j.patcog.2012.02.014
[25] Mahmood, N. et al., Blood cells extraction using color based segmentation technique, An International Journal IJLBPR, 2(2), 2013. DOI: 10.1016/j.patcog.2012.02.014
[26] Ramesh, N., Salama, M. and Tasdizen, T., Segmentation of haematopoeitic cells in bone marrow using circle detection and splitting techniques, 9th IEEE International Symposium on Biomedical Imaging (ISBI), 2012. pp. 206-209. DOI: 10.1109/ISBI.2012.6235520
[27] Otsu, N., A threshold selection method from gray-level histograms. Automatica, 11, pp. 23-27, 1975.
[28] Jayaraman. Digital image processing. Image segmentation. Nueva Delhi: Tata McGraw-Hill Education, 2011.
[29] Gomez, W., Leija, L., Alvarenga, A., Infantosi, A. and Pereira, W. Computerized lesion segmentation of breast ultrasound based on marker-controlled watershed transformation. Med Phys, 37, pp. 82-95, 2010. DOI: 10.1118/1.3265959
[30] Huang, J., An improved algorithm of overlapping cell division, Intelligent Computing and Integrated Systems (ICISS), 2010. pp. 687-691. DOI: 10.1109/ICISS.2010.5655507
[31] Chitade, A.Z. and Katiyar, S.K., Colour based image segmentation using k-means clustering. International Journal of Engineering Science and Technology 2(10), pp. 5319-5325, 2010.
[32] Vianney, J., Rosales, A. and Gallegos, F., Computer-aided diagnosis of brain tumors using image enhancement and fuzzy logic. DYNA, 81(183), pp. 148-157, 2014. DOI: 10.15446/dyna. v81n183.36838
M.F. Romero-Rondón, Received a BSc. Eng in Systems Engineering in 2014 from the Universidad Industrial de Santander, Bucaramanga, Colombia. He is currently a graduate student at the University of Nice-Sophia Antipolis, France. His current research interests include bioinformatics, biomedical signal and image analysis, telemedicine, and computer vision. ORCID: 0000-0001-6199-8722
L.M. Sanabria-Rosas, received a BSc. Eng in Systems Engineering in 2014 from the Universidad Industrial de Santander, Bucaramanga, Colombia. She has worked in the biomedicine and telemedicine fields. She is currently a student on the International Masters in Computer Science at the Université Nice-Sophia Antipolis. ORCID: 0000-0003-4345-1074
L.X. Bautista-Rozo, received a BSc. Eng. in Systems Engineering in 2003 from the Universidad Industrial de Santander, Bucaramanga, Colombia and an MSc. in Computer Engineering in 2007 from the University of Puerto Rico, Mayaguez Campus. She was the director of the Biomedical Engineering (GIIB) Research Group from 2011 to 2013. She is currently a PhD candidate at the Ecole Doctorale des Sciences et des Technologies de l'Information et de la Communication at the Université Nice-Sophia Antipolis. ORCID: 0000-0002-3853-007X
A. Mendoza-Castellanos, received a BSc. in Computer Science from New York Institute of Technology, an MSc in Automation, Robotics and Informatics from the Université Franche Comté. From 1980 to 2011 he was Full Professor in the Universidad Industrial de Santander. He was the founder and director of the Biomedical Engineering (GIIB) Research Group. He is currently the general manager of the company Biosys LTDA. ORCID: 0000-0002-3550-9638
Referencias
Rogers, K., Blood: Physiology and Circulation. New York: Britannica Educational, 2010
Bijlani, R., Fundamentals physiology a textbook for nursing students. New Delhi: Jaypee Brothers Publishers, 2001.
Estridge, B., Basic Medical Laboratory Techniques. Clifton Park: Cengage Learning, 2000.
Rodak, B., Hematology: Clinical principles and applications. Missouri: Elsevier Health Sciences, 2007.
Ruiz, G., Fundamentos de hematología. Buenos Aires: Ed. Médica Panamericana, 1994.
Kothari, S., Chaudry, Q. and Wang, M., Automated cell counting and cluster segmentation using concavity detection and ellipse fitting techniques, Biomedical Imaging: From Nano to Macro, 2009. pp. 795-798. DOI: 10.1109/ISBI.2009.5193169
Cloppet, F. and Boucher, A., Segmentation of complex nucleus configurations in biological images. Pattern Recognition Letters, 31(8), pp. 755-761, 2010. DOI: 10.1016/j.patrec.2010.01.022
Sharif, J., et al., Red blood cell segmentation using masking and watershed algorithm: A preliminary study, Biomedical Engineering (ICoBE), 2012. pp. 258-262. DOI: 10.1109/ICoBE.2012.6179016
Veta, M., et al., Marker-controlled watershed segmentation of nuclei in H&E stained breast cancer biopsy images, Biomedical Imaging: From Nano to Macro, 2011. pp. 618-621. DOI: 10.1109/ISBI.2011.5872483
Xu, S., Liu, H. and Song, E., Marker-controlled watershed for lesion segmentation in mammograms. Journal of Digital Imaging, 24(5), pp. 754-763, 2011. DOI: 10.1007/s10278-011-9365-2
Di Cataldo, S. et al.. Automated segmentation of tissue images for computerized IHC analysis. Computer Methods and Programs in Biomedicine, 100(1), pp. 1-15, 2010. DOI: 10.1016/j.cmpb.2010.02.002
Karvelis, P., Likas, A. and Fotiadis, D., Identifying touching and overlapping chromosomes using the watershed transform and gradient paths. Pattern Recognition Letters, 31(16), pp. 2474-2488, 2010. DOI: 10.1016/j.patrec.2010.08.002
Wen, Q., Chang, H. and Parvin, B., A Delaunay triangulation approach for segmenting clumps of nuclei, Biomedical Imaging: From Nano to Macro, 2009. pp. 9-12. DOI: 10.1109/ISBI.2009.5192970
Kumar, S. et al., A rule-based approach for robust clump splitting. Pattern Recognition, 39(6), pp. 1088-1098, 2006. DOI: 10.1016/j.patcog.2005.11.014
Wang, H., Zhang, H. and Ray, N., Clump splitting via bottleneck detection and shape classification. Pattern Recognition, 45(7), pp. 2780-2787, 2012. DOI: 10.1016/j.patcog.2011.12.020
Berge, H. et al., Improved red blood cell counting in thin blood smears, Biomedical Imaging: From Nano to Macro, 2011. pp. 204-207. DOI: 10.1109/ISBI.2011.5872388
Latorre, A. et al., Segmentation of neuronal nuclei based on clump splitting and a two-step binarization of images. Expert Systems with Applications, 40(16), pp. 6521-6530, 2013. DOI: 10.1016/j.eswa.2013.06.010
Di Ruberto, C., Dempster, A., Khan, S. and Jarra, B., Analysis of infected blood cell images using morphological operators. Image and Vision Computing, 20, pp. 133-146, 2002. DOI: 10.1016/S0262-8856(01)00092-0
Buggenthin, F. et al., An automatic method for robust and fast cell detection in bright field images from high-throughput microscopy. BMC Bioinformatics, 14(297), pp. 1-12, 2013. DOI: 10.1186/1471-2105-14-297
Prasad, K. et al., Image analysis approach for development of a decision support system for detection of malaria parasites in thin blood smear images. Journal of digital imaging, 25, pp. 542-549, 2012. DOI: 10.1007/s10278-011-9442-6
Amit-Kunar, P., Tembhare, P. and Pote, C., Enhanced identification of malarial infected objects using otsu algorithm from thin smear digital images. International Journal of Latest Research in Science and Technology. 1, pp. 159-163, 2012.
Priyankara, G.P.M. et al., An extensible computer vision application for blood cell recognition and analysis. Thesis, Department of Computer Science and Engineering, University of Moratuwa, Sri Lanka, 2006.
Guan, P.P. and Hong, Y., Blood cell image segmentation based on the Hough transform and fuzzy curve tracing. Machine Learning and Cybernetics (ICMLC), 2011. pp. 1696-1701. DOI: 10.1109/ICMLC.2011.6016961
Prasad, D.K., Leung, M.K. and Cho, S.Y., Edge curvature and convexity based ellipse detection method. Pattern Recognition, 45(9), pp. 3204-3221, 2012. DOI: 10.1016/j.patcog.2012.02.014
Mahmood, N. et al., Blood cells extraction using color based segmentation technique, An International Journal IJLBPR, 2(2), 2013. DOI: 10.1016/j.patcog.2012.02.014
Ramesh, N., Salama, M. and Tasdizen, T., Segmentation of haematopoeitic cells in bone marrow using circle detection and splitting techniques, 9th IEEE International Symposium on Biomedical Imaging (ISBI), 2012. pp. 206-209. DOI: 10.1109/ISBI.2012.6235520
Otsu, N., A threshold selection method from gray-level histograms. Automatica, 11, pp. 23-27, 1975.
Jayaraman. Digital image processing. Image segmentation. Nueva Delhi: Tata McGraw-Hill Education, 2011. Romero-Rondón et al / DYNA 83 (198), pp. 188-195, Septiembre, 2016. 195
Gomez, W., Leija, L., Alvarenga, A., Infantosi, A. and Pereira, W. Computerized lesion segmentation of breast ultrasound based on marker-controlled watershed transformation. Med Phys, 37, pp. 82-95, 2010. DOI: 10.1118/1.3265959
Huang, J., An improved algorithm of overlapping cell division, Intelligent Computing and Integrated Systems (ICISS), 2010. pp. 687-691. DOI: 10.1109/ICISS.2010.5655507
Chitade, A.Z. and Katiyar, S.K., Colour based image segmentation using k-means clustering. International Journal of Engineering Science and Technology 2(10), pp. 5319-5325, 2010.
Vianney, J., Rosales, A. and Gallegos, F., Computer-aided diagnosis of brain tumors using image enhancement and fuzzy logic. DYNA, 81(183), pp. 148-157, 2014. DOI: 10.15446/dyna. v81n183.36838
Cómo citar
IEEE
ACM
ACS
APA
ABNT
Chicago
Harvard
MLA
Turabian
Vancouver
Descargar cita
CrossRef Cited-by
1. Nataliya BİLOUS, Oleg HRAMM, İryna AHEKİAN, Abed Thamer KHUDHAIR, Ludmila ILLYASHENKO, Alexander NERUKH. (2019). NUMERICAL ERROR ANALYSIS FOR CONFIGURABLE CELL SEGMENTATION PROBLEM. Eskişehir Technical University Journal of Science and Technology A - Applied Sciences and Engineering, 20, p.193. https://doi.org/10.18038/estubtda.650048.
2. Sapto Nisworo, Alifia Revan Prananda. (2023). A Review on Digital Microscopic Images for Plasmodium Parasite Detection. TEKNIK, 44(3), p.222. https://doi.org/10.14710/teknik.v44i3.53990.
3. Asaad Ma. Babker, Rania Saad Suliman, Rabab Hassan Elshaikh, Sardor Boboyorov, Vyacheslav Lyashenko. (2024). Sequence of Simple Digital Technologies for Detection of Platelets in Medical Images. Biomedical and Pharmacology Journal, 17(1), p.141. https://doi.org/10.13005/bpj/2842.
4. Fatih Veysel Nurçin. (2022). Improved segmentation of overlapping red blood cells on malaria blood smear images with TransUNet architecture. International Journal of Imaging Systems and Technology, 32(5), p.1673. https://doi.org/10.1002/ima.22739.
5. Prasenjit Dhar, K. Suganya Devi, Satish Kumar Satti, P. Srinivasan. (2025). Efficient detection and partitioning of overlapped red blood cells using image processing approach. Innovations in Systems and Software Engineering, 21(1), p.79. https://doi.org/10.1007/s11334-022-00478-y.
6. Fatih Veysel Nurçin, Elbrus Imanov, Andrea Picone. (2021). Selective Hole Filling of Red Blood Cells for Improved Marker-Controlled Watershed Segmentation. Scanning, 2021, p.1. https://doi.org/10.1155/2021/5678117.
7. Mohamed Ait Mehdi, Khadidja Belattar, Feriel Souami. (2023). Artificial Intelligence Doctoral Symposium. Communications in Computer and Information Science. 1852, p.95. https://doi.org/10.1007/978-981-99-4484-2_8.
Dimensions
PlumX
Visitas a la página del resumen del artículo
Descargas
Licencia
Derechos de autor 2016 DYNA

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
El autor o autores de un artículo aceptado para publicación en cualquiera de las revistas editadas por la facultad de Minas cederán la totalidad de los derechos patrimoniales a la Universidad Nacional de Colombia de manera gratuita, dentro de los cuáles se incluyen: el derecho a editar, publicar, reproducir y distribuir tanto en medios impresos como digitales, además de incluir en artículo en índices internacionales y/o bases de datos, de igual manera, se faculta a la editorial para utilizar las imágenes, tablas y/o cualquier material gráfico presentado en el artículo para el diseño de carátulas o posters de la misma revista.