Detection and sequencing of Potato virus Y (PVY) and Potato leafroll virus (PLRV) in a volunteer plant of Solanum tuberosum L. cv. Diacol-Capiro
Detección y secuenciación del Potato virus Y (PVY) y Potato leafroll virus (PLRV) a partir de una planta voluntaria de Solanum tuberosum L. cv. Diacol-Capiro
DOI:
https://doi.org/10.15446/acag.v66n4.59753Palabras clave:
Polerovirus, Potyvirus, RT-qPCR, High-throughput sequencing, Solanaceae (en)Polerovirus, Potyvirus, RT-qPCR, Secuenciación masiva de nueva generación, Solanaceae (es)
Descargas
Viral diseases are among the most limiting factors in the production of potato in Colombia and the rest of the world. The best strategy to control plant viruses consists on the use of certified seed tubers, control of arthropod vectors and the use of adequate crop management practices that reduce mechanical transmission and the presence of viral reservoirs like weeds and volun-teer plants. However, the successful implementation of these practices relies on the availability of highly sensitive techniques that allow for the asymptomatic detection of viruses. In this work, we tested the performance of Next-generation sequencing (NGS) and real time RT-PCR (RT-qPCR) on a single volunteer potato plant (cv. Diacol-Capiro) growing naturally in a seed-tuber storage facility in Yarumal (Antioquia). Our NGS results demonstrate a mixed infection with Potato virus Y (PVY) and Potato leafroll virus (PLRV). RT-qPCR was performed in roots, main stolons, crown (root collar) and upper, middle and lower leaves using specific primers for PVY, PLRV, Potato virus S (PVS), Potato virus V (PVV), Potato virus X (PVX) and Potato yellow vein virus (PYVV). Only PVY and PLRV were detected in good agreement with the NGS data. This work demonstrates the use-fulness of both techniques for supporting integrated management of plant viruses in potato, in-cluding virus detection in natural reservoirs such as volunteer plants and weeds.
Recibido: 24 de agosto de 2016; Aceptado: 5 de abril de 2017
Abstract
Viral diseases are among the most limiting factors in the production of potato in Colombia and the rest of the world. The best strategy to control plant viruses consists on the use of certified seed tubers, control of arthropod vectors and the use of adequate crop management practices that reduce mechanical transmission and the presence of viral reservoirs like weeds and volunteer plants. However, the successful implementation of these practices relies on the availability of highly sensitive techniques that allow for the asymptomatic detection of viruses. In this work, we tested the performance of Next-generation sequencing (NGS) and real time RT-PCR (RT-qPCR) on a single volunteer potato plant (cv. Diacol-Capiro) growing naturally in a seed-tuber storage facility in Yarumal (Antioquia). Our NGS results demonstrate a mixed infection with Potato virus Y (PVY) and Potato leafroll virus (PLRV). RT-qPCR was performed in roots, main stolons, crown (root collar) and upper, middle and lower leaves using specific primers for PVY, PLRV, Potato virus S (PVS), Potato virus V (PVV), Potato virus X (PVX) and Potato yellow vein virus (PYVV). Only PVY and PLRV were detected in good agreement with the NGS data. This work demonstrates the usefulness of both techniques for supporting integrated management of plant viruses in potato, including virus detection in natural reservoirs such as volunteer plants and weeds.
Keywords:
Polerovirus, Potyvirus, RT-qPCR, High-throughput sequencing, Solanaceae.Resumen
Las enfermedades virales son uno de los principales limitantes para la producción de papa en Colombia y otros países. El manejo de los virus de plantas se fundamenta en la siembra de material certificado por su sanidad, el control de vectores artrópodos y la realización de prácticas culturales que eviten su transmisión mecánica y mantenimiento en reservorios naturales como plantas voluntarias y arvenses. Sin embargo, el éxito de estas prácticas requiere de la posibilidad de disponer de técnicas altamente sensibles para la detección asintomática de virus. En este trabajo se evaluaron dos de dichas metodologías, como son la secuenciación masiva de nueva generación (NGS) y la RT-PCR en tiempo real (RT-qPCR), sobre una planta voluntaria de papa cv. Diacol-Capiro que crecía en la calle de un silo rústico de almacenamiento de tubérculos-semilla en Yarumal (Antioquia). Los resultados indicaron la infección mixta de la planta por Potato virus Y (PVY) y Potato leafroll virus (PLRV). Las pruebas de RT-qPCR con cebadores específicos para ambos virus y para Potato virus S (PVS), Potato virus V (PVV), Potato virus X (PVX) y Potato yellow vein virus (PYVV), confirmaron la infección de PVY y PLRV en seis tejidos diferentes (raíz, estolón principal, cuello y tejido foliar de tercios inferior, medio y superior), pero no de los otros virus. Este trabajo demuestra la utilidad de ambas técnicas moleculares para apoyar los programas de manejo integrado de enfermedades virales en papa, incluyendo la detección de reservorios naturales como plantas voluntarias y arvenses.
Palabras clave:
Polerovirus, Potyvirus, RT-qPCR, Secuenciación masiva de nueva generación, Solanaceae.Introduction
The potato (Solanum tuberosum L. and Solanum phureja Juz. & Bukasov) crop is one of the most important agricultural sectors in the Andean regions of Colombia comprising about 144097 ha for 2014, concentrated in the provinces of Cundinamarca (50480 ha), Boyacá (44993 ha), Nariño (30782 ha), Antioquia (7065 ha) and Norte de Santander (3910 ha), with a national annual production of 2758241 t.ha-1 (http://agronet.gov.co). In spite of the Andes being the center of origin of potato, the estimated yield for 2015 was only 19.9 t.ha-1, which is far below the yields reported for developed countries such as the United States (51.7 t.ha-1), the Netherlands (50.3 t.ha-1), Canada (36.4 t.ha-1) and the United Kingdom (33.17 t.ha-1) (http://faostat3.fao.org). As a result of using uncertified seed-tubers and poor management of arthropod vectors, viral diseases are one of the most limiting factors of productivity in potato crops (Halterman, Charkowski & Verchot, 2012). Colombia has a demanding potato certification program monitored by ICA that forbids the presence of Potato virus Y (PVY), Potato leafroll virus (PLRV), Potato virus S (PVS) and Potato virus X (PVX) in the Super Elite and Elite materials used to start the seed production cycle and levels of infection below 1% (basic), 2% (registered) and 5% (certified) for tuber-seeds obtained from the field (http://www.ica.gov.co). However, in Colombia there are serious deficiencies in the use of certified plant material, since it is estimated that only 3% of the planted tuber-seeds are certified, most of which correspond to the Diacol-Capiro variety (90%) (http://sioc.minagricultura.gov.co). Besides PVY, PLRV, PVS, PVX and PYVV, there are at least other 35 virus species that affect potato crops (Kerlan, 2008). For example, Halterman, Charkowski & Verchot (2012), identified PVY, PLRV, Potato virus A (PVA), Potato virus M (PVM), Tobacco rattle virus (TRV) and Potato mop top virus (PMTV) as the most limiting viruses in North America as they affect the S. tuberosum subsp. tuberosum varieties cultivated in the temperate region; while Andean potato latent virus (APLV), PYVV, Wilt potato mosaic virus (WPMV) and Potato yellowing virus (PYV) are another prevalent viruses in the South American Andes (Kerlan, 2008).
In recent recent years, metagenomic studies using Next-generation sequencing (NGS) methods have greatly increased our knowledge on viruses infecting a wide range of hosts including plants (Liu, Li, Li, Hu, He, Pong, Lin, Lu & Law, 2012; Ho & Tzanetakis, 2014). With these methods, it is possible to identify and characterize pathogens at the molecular level without any previous sequence knowledge and due to the high volume of data generated (1 Gb-100 Gb), NGS has a dynamic range that allow detection of sequences with very low abundance (Liu et al., 2012). This is in marked contrast with the Sanger sequencing method that have a maximum output in an ABI 3730xl analyzer of about 64 kb per run (Liu et al., 2012). With the use of NGS in the study of potato crops in Colombia it has been possible to detect and characterize either the complete or partial genome sequences of APLV (Kreuze, Koenig, de Souza, Vetten, Muller, Flores, Ziebell & Cuellar, 2013); PVY (Villamil, Cuellar & Guzmán, 2014; Muñoz, Gutiérrez & Marín, 2016), PVS (Gutiérrez, Alzate & Marín, 2013), PVV (Gutiérrez, Jaramillo & Marín, 2016) and PYVV (Villamil, Cuellar & Guzmán, 2014; Álvarez, Gutiérrez & Marín, 2017).
During an investigation on the phytosanitary status of commercial tuber-seeds in different potato-growing regions of Antioquia, we noticed a rustic storage silo of S. tuberosum subsp. andigena cv. Diacol-Capiro with volunteer plants growing in the corridors, exhibiting symptoms typical of viral infection and highly infested by aphids. These facts are obvious evidence on the lack of knowledge about virus reservoirs and transmission mechanisms in local farmers. The aim of this work, was to evaluate the usefulness of the Illumina NGS HiSeq 2000 platform and RT-qPCR as virus detection tools in different potato tissues and the potential use of these techniques as standard procedures in supporting integrated virus disease management programs in this crop.
Materials and methods
Plant material
This study was performed on a single plant of S. tuberosum subsp. andigena cv. Diacol-Capiro collected at Llanos de Cuivá (Yarumal, Antioquia). This plant was growing naturally in a central corridor within a rustic tuber-seed storage silo, exhibited leaf yellowing symptoms and was completely infested with aphids (Figure 1).
Figure 1: Condition of the rustic tuber-seed storage silo at Llanos de Cuivá (Yarumal, Antioquia) (A, B, C) where the volunteer potato plant used in this study was collected. A detail of the plant used in this study showing a high infestation by aphids (D).
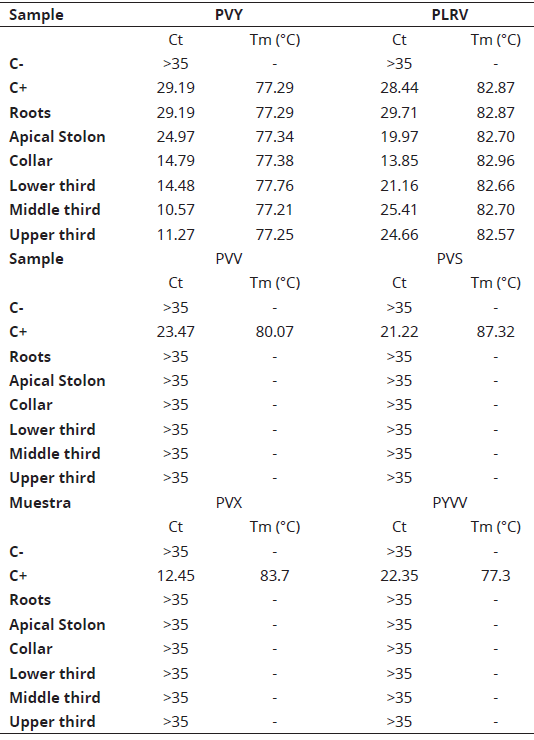
Six tissue sections from roots, the main stolon, crown (root collar) and leaves from the lower, middle and upper thirds were extracted with a sterile scalpel to be used in the molecular detection of viruses (Table 1).
Table 1: RT-qPCR detection of RNA viruses in six different tissues of a volunteer potato plant collected in a storage silo at Llanos de Cuivá (Yarumal, Antioquia).
Next-generation sequencing
Tissue samples were grounded separately using liquid nitrogen and total RNA extracted with the GeneJET Plant RNA Purification Mini kit (Thermo, EEUU); a composite sample containing 2 µl of total RNA from each tissue (12 µl in total) was made. Ribosomal RNA was depleted with the kit Ribo-Zero rRNA Removal kit (Illumina, EEUU) and the RNA integrity number (RIN) measured in a 2100 Bioanalyzer (Agilent Technologies, EEUU). The cDNA library was constructed with the TruSeq RNA Sample Preparation kit (Illumina, EEUU) and sequencing performed in an Illumina HiSeq 2000 equipment at Macrogen (South Korea).
Bioinformatic analyses
After NGS, low quality bases (Phred <30) were removed from the data set using SeqTK (https://github.com/lh3/seqtk). Viral sequences were identified using BLASTN and BLASTX (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against a local database containing all viruses infecting plants in the Ninth Report of the International Committee of Taxonomy of Viruses (King, Adams, Carstens & Lefkowitz, 2012) and Viralzone (http://viralzone.expasy.org). Viral genomes were assembled by mapping to reference genomes using Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Assemblies were verified for inconsistencies and sequencing errors with Tablet (https://ics.hutton.ac.uk/tablet). Open reading frames (ORFs) were identified with BLASTX (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Consensus sequences for PVY and PLRV were deposited in GenBank under accession codes KX756672 and KX712226, respectively.
Phylogenetic analyses of complete genomes were performed in MEGA6 by the Maximum-likelihood (ML) method using the Tamura-3 substitution model and 1000 bootstrap replicates (http://www.megasoftware.net). In each case, a representative set of sequences reflecting the diversity of strains and geographical distribution was chosen. Sequence alignments were performed with MUSCLE, gaps and missing data were removed when their site coverage was superior to 95%. Rate variation among sites was estimated using a gamma distribution. The initial tree for the ML method was obtained using the Neighbor-Joining method and the likelihood improved by the Nearest-Neighbor-Interchange heuristic. Phylogenetic relationships are presented as mid-point rooted trees with branch lengths proportional to the number of base substitutions per site.
RT-qPCR
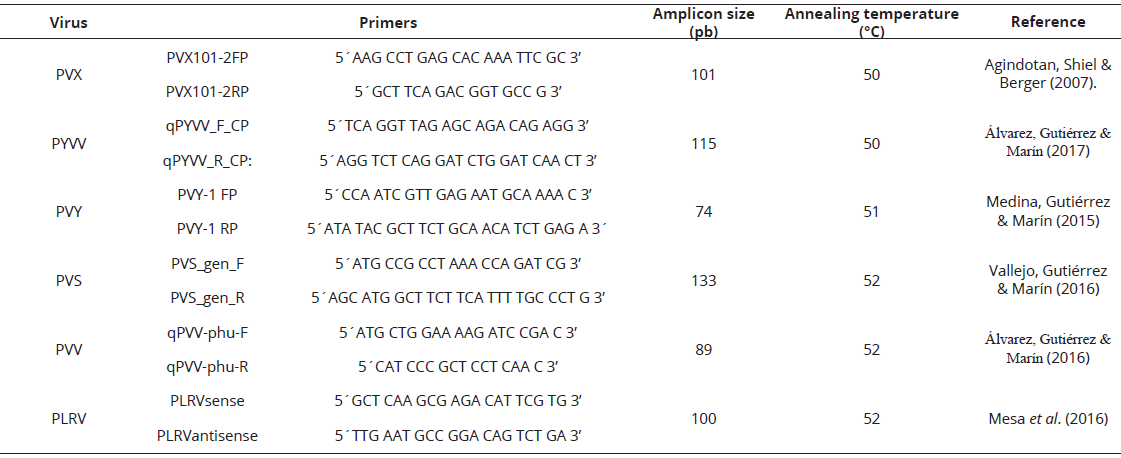
NGS results were confirmed by a two-step RT-qPCR method using SYBR Green I. Each of the six tissue samples was tested for PVY, PLRV, PVX, PVV, PVS and PYVV, all of which have been reported to infect potato in Colombia. All primers targeted the capsid region of each virus and have been reported in previous studies; details are shown in Table 2. Retrotranscription was performed in a 20 µL reaction volumen containing 200 units of Maxima Reverse Transcriptase (Thermo, EEUU), 1X RT buffer, 0.5 mM dNTPs, 20 units RiboLock RNAse Inhibitor (Thermo, EEUU) and 4 μL of total RNA. For PVY, PVV and PVS, 20 pmoles of Oligo-(dT)18 were used as reverse primer; while a specific reverse primer was used for PVX, PYVV and PLRV. Reactions were incubated in a T3 thermocycler (Biometra, Germany) at 65°C for 5 min, followed 50°C for 30 min and a final step at 85°C for 5 min. qPCR were performed in a 20 µL reaction volumen containing 12.5 µL of Maxima SYBR Green/ROX qPCR Master Mix (2X) (Thermo, EEUU), 10 µL DEPC-treated water, 0.3 µM of each primer and 50 ng of cDNA. The amplification reaction was performed in a Rotor-Gene Q-5plex Platform (Qiagen, Germany) and consisted of a Taq-polymerase activation step at 95°C for 10 min and 35 cycles at 95°C for 10 s and 45 s at corresponding annealing temperature (Table 2).
Table 2: Primer used for RT-qPCR detection of RNA viruses in this study.
Fluorescense was measured after each amplification cycle. Ct values were determined using the default parameters of the Rotor-Gene Q 1.7 software. Samples were considered positive when they reached the Ct value before the 35th cycle (Schena, Nigro, Ippolito & Gallitelli, 2004). A High Resolution Melting (HRM) analysis was performed for each sample in the 50-99°C range and Tm values compared with the positive control for each virus. All tests included a negative control consisting of a sample lacking viral cDNA.
Results
Next-Generation sequencing
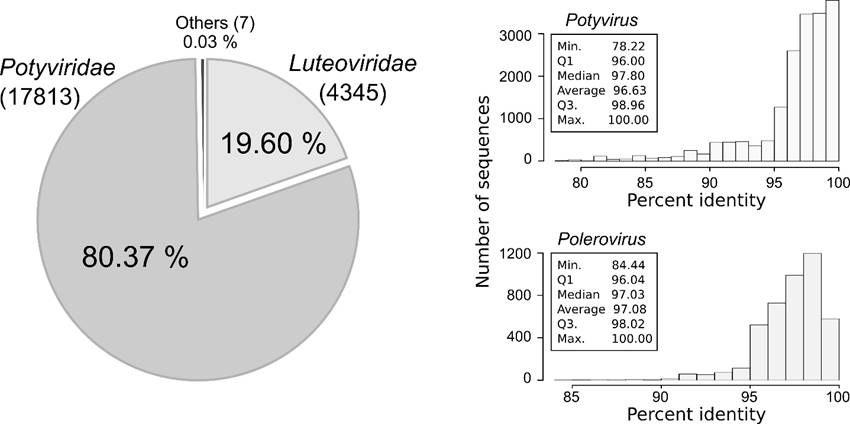
NGS transcriptome sequencing of the potato cv. Diacol-Capiro volunteer plant resulted in a paired-end library of 6980917 reads for a total of 1396183400 nt. BLAST analyses identified 17813 and 4345 reads with significant similarity to plant viruses. Potyviridae (80.37%) and Luteoviridae (19.6%) accounted for the majority of putative viral sequences in the transcriptome; only seven reads (0.03%) were associated to other virus taxa (Figure 2).
Figure 2: NGS detection of RNA viruses in a volunteer potato plant within a seed-tuber storage silo in Llanos de Cuivá (Yarumal, Antioquia). The total number and percentage of reads with significant sequence similarity to plant virus families are shown to the left. At the genus level, viral reads were more closely related to viruses in the genus Potyvirus and Polerovirus. Histograms showing the distribution of sequence identiy is shown to the right.
Analysis at the genus levels indicated that the viral sequences in the transcriptome are highly similar to viruses within the genus Potyvirus and Polerovirus with average percent nucleotide identities of 96.63% and 97.08%, respectively (Figure 2). The virus species were identified as PVY (Potyvirus) and PLRV (Polerovirus) and their complete genomes were assembled with a coverage of 701.8x and 160x, respectively.
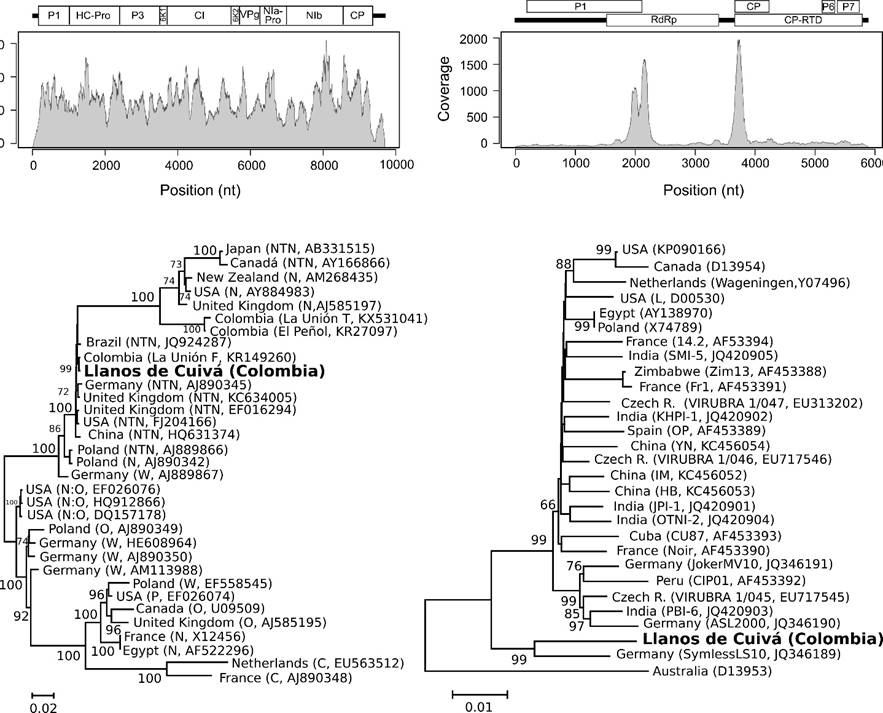
A phylogenetic analysis of complete PVY genomes revealed the existence of four main clades (100% bootstrap) that correspond to the main PVY lineages: Necrotic (N/NTN), Necrotic recombinant (N:O; W), Ordinary (O) and stipple streak (C), (Figure 3).
RT-qPCR
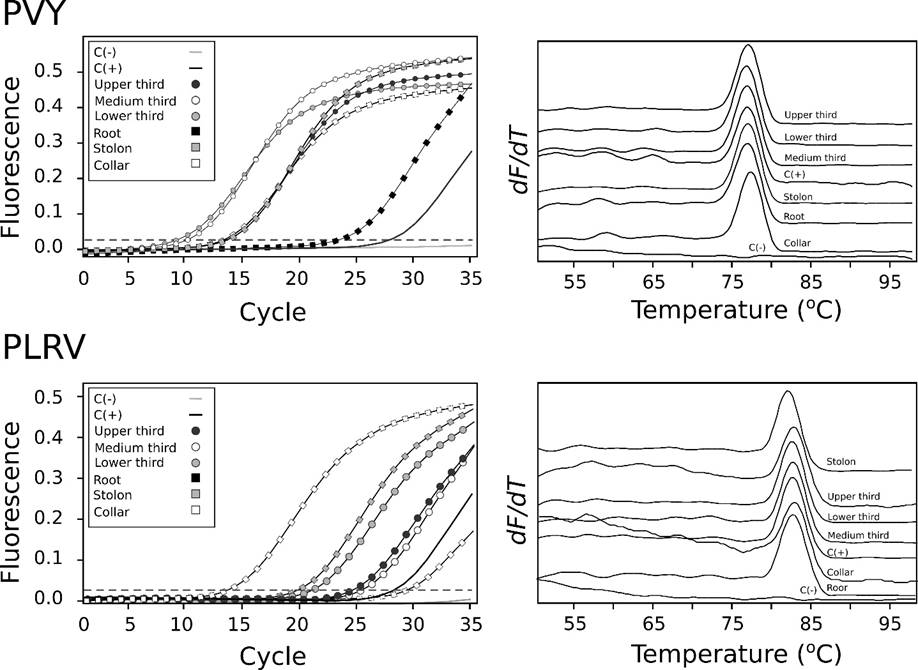
Infected samples with each virus were used as positive controls. Our results suggest the presence of both PVY and PLRV in all the tissue samples analyzed (Figure 4); Ct values were in the 10.57 to 29.19 range for PVY and between 13.85 and 29.71 for PLRV. The highest titer was found to occur in the leaf tissue (Ct = 10.57-14.48) for PVY and the collar root for PLRV (Ct = 13.85) (Table 1). As the Ct value is inversely proportional to the concentration of template nucleic acids, foliar tissue should be used as source material for PVY detection.
Figure 3: Sequence coverage and annotation of the PVY (top left) and PLRV (top right) genomes assembled from the transcriptome data. Maximum-likelihood phylogenetic trees for the PVY (botton left) and PLRV (bottom right) genomes. For PVY, sequences are a representative set of strains reflecting the diversity of strains and geographical distribution of this virus; PLRV sequences include all currently available complete genomes for this virus in GenBank. Tree relationships are presented as mid-point rooted trees with branch lengths proportional to the number of base substitutions per site.
Figure 4: RT-qPCR amplification curves (Left) for PVY and PLRV in six different tissues of a volunteer potato plant collected in a storage silo at Llanos de Cuivá (Yarumal, Antioquia). SYBR-Green I was used as fluorescent dye. The melting curve profiles for each sample are shown to the right.
Discussion
The PVY genome comprised 9,704 nucleotides (excluding the poly(A) tail), with 5' and 3' UTR regions of 190 and 331 nt, respectively. The PVY genome contains a single ORF encoding a protein of 3,061 amino acid, which is expected to be proteolitically processed into ten mature protein products (King, Adams, Carstens & Lefkowitz, 2012): P1, HC-Pro, P3, PIPO, 6K1, CI, 6K2, VPg, NIa-Pro, NIb and CP (Figure 3). The sequence motif GA7T giving rise to frameshift protein product P3N-PIPO (243 a.a.) was identified at nucleotide position 2921-2929 within the P3 segment. A conserved potyviral GDD motif was found at amino acid positions 2626-2628 within the RdRP; while the DAG sequence involved in aphid transmission was detected between residues 2800-2802 near the CP N-terminus (Revers & García, 2015).
The PVY isolate from Llanos de Cuivá clustered within the PVYN/NTN clade in a subgroup that also includes isolate PVY_La UniónF (KR149260), reported by Muñoz, Gutiérrez & Marín (2016), in a NGS study of leaf tissues from potato cv. Diacol-Capiro. This result is also in good agreement with a previous study by Henao, Gutiérrez, & Marín (2013), where, using partial CP sequencing, the authors showed the widespread presence of PVY genotypes closely related to strains in the PVYN/NTN group; similar results have been obtained for PVY strains in the province of Cundinamarca using NGS of miRNAs and Sanger sequencing of the CP segment (Villamil, Cuellar & Guzmán, 2014).
To our knowledge, this is the first complete PLRV genome from Colombia and the second from South America after a report by Guyader & Ducray (2002), in Perú (AF453392). Phylogenetic analysis of complete genomes showed a majoritary (26 sequences) and a minoritary (two sequences) clade with strong bootstrap support (99%). The PLRV isolate from Llanos de Cuivá clustered in the minoritary clade together with strain SymlessS10 (JQ346189) from Germany (Figure 3). Isolate SymlessS10 is a low-pathogenic strain that do not induce visible symptoms; it would be interesting to perform pathogenicity tests for the PLRV isolate from Llanos de Cuivá on the potato varieties cultivated in Colombia to determine if these genotypes have hypovirulent characteristics.
With respect to PLRV, the genome comprised 5881 nt with 5' and 3' UTR regions of 77 and 405 nt, respectively (Figure 3). The following ORFs typical of PLRV were found: supressor of silencing P0 (ORF0), protein P1 involved in replication and proteolytic processing of VPg (ORF1), the viral RNA-dependent RNA polymerase (RdRP, ORF2), the major capsid protein (ORF3), a movement protein (ORF4), a 80 kDa readthrough protein product involved in aphid transmission (ORF3/ORF5), a 6 kDa protein of unknown function (ORF6), nucleic acid binding protein P7 (ORF7) and protein Rap1 involved in virus replicacion (ORF8) (Guyader & Ducray, 2002).
Due to differences in sensitity and experimental approach, it is typical in virological studies to combine biological, serological and molecular methods to cofirm diagnostic results (Kerlan, 2008; Wang, Ma, Zhang, Wu, Wu, Wang & Li, 2011). For this reason, NGS results were confirmed by RT-qPCR. The primers used in this study have been previously demostrated to be effective in detecting Colombian strains of PVY and PLRV (Medina, Gutiérrez & Marín, 2015; Mesa, González, Gutiérrez & Marín, 2016). Samples were also tested for the presence of other viruses infecting potato in Colombia: PVX (García, Olarte, Gutiérrez & Marín, 2016), PYVV (Álvarez, Gutiérrez & Marín, 2017), PVS (Vallejo, Gutiérrez & Marín, 2016) and PVV (Gutiérrez et al., 2016).
With respect to PLRV, vascular tissue would be the preferred source material, which is consistent with phloem-restriction of this virus (Guyader & Ducray, 2002). Melting temperature values of the amplicons was in good agreement with previous reports by Medina, Gutiérrez & Marín (2015), for PVY (Tm=77.5°C) and Mesa, González, Gutiérrez & Marín (2016), for PLRV (Tm=81.8°C). With the exception of the positive controls, all samples tested negative for PVX, PYVV, PVS and PVV (Table 1). The RT-qPCR results match well with the NGS data, validating its potential as the molecular diagnostic tool of choice in the asymptomatic detection of known viruses, since it has been estimated that RT-qPCR is about 103-105 times more sensitive than either ELISA or conventional RT-PCR (Bertolini, Moreno, Capote, Olmos, de Luis, Vidal, Pérez-Panadés & Cambra, 2008; Medina, Gutiérrez & Marín, 2015).
The integrated managment of viral diseases in potato are highly dependent on the use of certified virus-free seeds and should be complemented with other sanitary measures, which in the production of tuber seeds are: avoidance of continuous cropping systems, elimination of alternate hosts such as weeds and volunteer plants, visual inspection of symptoms and removal of infection foci, use of highly sensitive detection methods in random samples to discard the presence of infected asymptomatic plants, aplication of mineral oils to reduce the dispersion of aphids and restrict agricultural practices that facilitate mechanical transmission (Coutts & Jones, 2015).
The tuber-seed certification process starts with the cleaning and propagation of a nuclear stock plant material using tissue culture techniques, which results in the production of mini-tubers that are further propagated under controlled greenhouse conditions. The tubers produced at this stage are multiplied in the field by tuber-seed producing farmers, which are under constant monitoring and surveillance by the corresponding governmental agencies (Halterman, Charkowski & Verchot, 2012).
The usefulness of NGS as a virus diagnostic tool in tuber-seed certification programs is currently limited by its high cost ((1000 USD per sample/3 Gb) but could probably be used in the inspection of the in vitro micro-plants due to its high sensitivity and usefulness in detecting novel variant or virus species. Recently, Ho & Tzanetakis (2014), showed that the NGS bioinformatic analysis can detect even a single read of viral origin in a database containing 25 to 30 million sequences. In addition, the RT-qPCR technique could be used as a diagnostic tool during the field stage of tuber-seed certification programs as its cost per sample (( 20 USD) is affordable for seed companies and governmental phytosanitary agencies; furthermore, this method only requires of a basic nucleic acid extraction, is fast (2-3 hours) and easy to implement in any laboratory.
The use of NGS and RT-qPCR in the Colombian agroindustry is urgently required as support in certifying the sanity of the nuclear stock plants, as the in vitro propagation methods does not guaranteed the obtention of virus free plants. For example, Wang et al. (2011), analyzed in vitro potato plants in the province of Schezuan (China) and detected infection by PVM, PVS, PVX and PVY in about 50% of the samples; the same analysis on the southern and northern regions found significant levels of infection by PLRV (up to 3%), PVS (up to 20.7%), PVX (up to 17%) and PVY (up to 18%).
In this work, we have shown infection by PVY and PLRV in a single volunteer plant growing within a silo that stored certified tuber-seed together with recently harvested tubers; several volunteer plants infested with aphids and exhibiting symptoms of viral infection (i.e. mosaics and leaf yellowing) were also observed in the premises. This study highlights the poor implementation of phytosanitary measures by some potato farmers such as inadequate handling of tuber-seeds, failure to eradicate volunteer plants and poor vector-control practices. Correcting these widespread malpractices is a requirement to improve the yield and environmental sustainability of this important crop in Colombia.
Conclusions
Using Next-generation sequencing, the complete genome of PVY (9,704 nt) and PLRV (5,881 nt) isolates infecting a single volunteer plant of potato cv Diacol-Capiro growing in the corridor of a rusitc tuber-seed storage facility in los Llanos de Cuivá (Yarumal, Antioquia) were obtained. Phylogenetic analyses showed the PVY isolate to be related to the necrotic strains N/NTN, while the PLRV isolate formed an independent clade together with a German isolate shown to exhibit low virulence levels in the field. The PLRV sequence presented here constitutes the first complete genome report from a Colombian isolate of this virus and the second in South America.
The monitoring of the phytosanitary status of potato crops in Colombia and other Andean countries with highly sensible techniques such as NGS and RT-qPCR must be a short-term target for this agroindustry. Given these concerns, this is particularly important in the diagnosis of viruses at different stages in the certification processs of tuber-seeds.
Acknowledgments
This work was financed by Vicerrectoría de Investigaciones, Universidad Nacional de Colombia (Grant: 34585).
References
Referencias
Agindotan, B.O., Shiel, P.J. & Berger, P.H. (2007). Simultaneous detection of potato viruses, PLRV, PVA, PVX and PVY from dormant potato tubers by TaqMan real-time RT-PCR. J Virol Methods, 142(1-2), 1-9. http://dx.doi.org/10.1016/j.jviromet.2006.12.012
Álvarez, D., Gutiérrez, P. & Marín, M. (2016). Caracterización molecular del Po-tato virus V (PVV) infectando Solanum phureja mediante secuenciación de nueva generación. Acta Biol Colomb, 21(3), 521-531. http://dx.doi.org/10.15446/abc.v21n3.54712
Álvarez, D., Gutiérrez, P. & Marín, M. (2017). Secuenciación del genoma del Po-tato yellow vein virus (PYVV) y desarrollo de una prueba molecular para su detección. Bioagro, 29(1), 3-14. http://www.scielo.org.ve/pdf/ba/v29n1/art01.pdf.
Bertolini, E., Moreno, A., Capote, N., Olmos, A, de Luis, A., Vidal, E., Pérez-Panadés, J. & Cambra, M. (2008). Quantitative detection of Citrus tristeza virus in plant tissues and single aphids by Real-time RT-PCR. Eur J Plant Pathol, 120(2), 177-188. http://dx.doi.org/10.1007/s10658-007-9206-9
Coutts, B.A. & Jones, R.A.C. (2015). Potato virus Y: Contact transmission, sta-bility, inactivation, and infection sources. Plant Dis, 99(3), 387-394. http://dx.doi.org/10.1094/PDIS-07-14-0674-RE
García, D., Olarte, M., Gutiérrez, P. & Marín, M. (2016). Detección serológica y molecular del Potato virus X (PVX) en tubérculos-semilla de papa (Sola-num tuberosum L. y Solanum phureja Juz. & Bukasov) en Antioquia, Co-lombia. Rev Colomb Biotecn, 18(1), 104-111. http://dx.doi.org/10.15446/rev.colomb.biote.v18n1.51389
Gutiérrez, P.A., Alzate, J.F. & Marín, M. (2013). Complete genome sequence of a novel Potato virus S strain infecting Solanum phureja in Colombia. Arch Virol, 158(10), 2205-2208. http://dx.doi.org/10.1007/s00705-013-1730-7
Gutiérrez, P.A., Jaramillo, H. & Marín, M. (2016). Genome sequence of a diver-gent Colombian isolate of Potato virus V (PVV) infecting Solanum phureja. Acta Virol, 60(1), 49-54. http://dx.doi.org/10.4149/av_2016_01_49
Guyader, S. & Ducray, G.D. (2002). Sequence analysis of Potato leafroll virus isolates reveals genetic stability, major evolutionary events and differential selection pressure between overlapping reading frame products. J Gen Vi-rol, 83(7), 1799–1807. http://dx.doi.org/10.1099/0022-1317-83-7-1799
Halterman, D., Charkowski, A. & Verchot, J. (2012). Potato, viruses, and seed certification in the USA to provide healthy propagated tubers. Pest Tech, 6(1), 1-14. http://www.globalsciencebooks.info/Online/GSBOnline/images/2012/PT_6(SI1)/PT_6(SI1)1-14o.pdf.
Henao, E., Gutiérrez, P. & Marín, M. (2013). Análisis filogenético de aislamien-tos del Potato virus Y (PVY) obtenidos en cultivos de papa (Solanum tube-rosum) y tomate de árbol (Solanum betaceum) en Colombia. Actual Biol, 35(99), 219-232. http://www.scielo.org.co/pdf/acbi/v35n99/v35n99a8.pdf.
Ho, T. & Tzanetakis, I.E. (2014). Development of a virus detection and discovery pipeline using next generation sequencing. Virology, 471-473, 54-60. http://dx.doi.org/10.1016/j.virol.2014.09.019
Kerlan, C. (2008). Potato viruses. Desk encyclopedia of plant and fungal virolo-gy In: B.W. Mahy & M.H. van Regenmortel (Eds.). Academic Press, Oxford, United Kingdom. 471 p.
King, A.M.Q., Adams, M.J., Carstens, E.B. & Lefkowitz, E.J. (2012). Virus tax-onomy: classification and nomenclature of viruses. Ninth Report of the In-ternational Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, EEUU. 1327 p.
Kreuze, J., Koenig, R., De Souza, J., Vetten, H.J., Muller, G., Flores, B., Ziebell, H. & Cuellar, W. (2013). The complete genome sequences of a Peruvian and a Colombian isolate of Andean potato latent virus and partial se-quences of further isolates suggest the existence of two distinct potato-infecting tymovirus species. Virus Res, 173(2), 431-435. http://dx.doi.org/10.1016/j.virusres.2013.01.014
Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., Lin, D., Lu, L. & Law, M. (2012). Comparison of next-generation sequencing systems. J Biomed Biotechnol, 2012, 1-12. http://dx.doi.org/10.1155/2012/251364
Medina, H.C., Gutiérrez, P. & Marín, M. (2015). Detección del Potato virus Y (PVY) en tubérculos de papa mediante TAS-ELISA y qRT-PCR en Antio-quia (Colombia). Bioagro, 27(2), 83-92. http://www.scielo.org.ve/pdf/ba/v27n2/art04.pdf.
Mesa, M., González, M., Gutiérrez, P. & Marín, M. (2016). Diagnóstico serológi-co y molecular del Potato leafroll virus (PLRV) en tubérculos-semilla de papa en Antioquia, Colombia. Acta Agron, 65(2), 204-210. http://dx.doi.org/ 10.15446/acag.v65n2.50764
Muñoz, D., Gutiérrez, P. & Marín, M. (2016). Detección y caracterización mole-cular del Potato virus Y (PVY) en cultivos de papa (Solanum tuberosum L.) del norte de Antioquia, Colombia. Rev Prot Veg, 31(1), 1-11. http://scielo.sld.cu/pdf/rpv/v31n1/rpv02116.pdf.
Revers, F. & García, J.A. (2015). Molecular biology of potyviruses. Adv Virus Res, 92, 101-199. http://dx.doi.org/10.1016/bs.aivir.2014.11.006
Schena, L., Nigro, F., Ippolito, A. & Gallitelli, D. (2004). Real-time quantitative PCR: a new technology to detect and study phytopathogenic and antago-nistic fungi. Eur J Plant Pathol, 110(9), 893-908. http://dx.doi.org/10.1007/s10658-004-4842-9
Vallejo, D., Gutiérrez, P. & Marín, M. (2016). Genome characterization of a Po-tato virus S (PVS) variant from tuber sprouts of Solanum phureja Juz. et Buk. Agron Colomb, 34(1), 51-60. http://dx.doi.org/10.15446/agron.colomb.v34n1.53161
Villamil-Garzón, A., Cuellar, W.J. & Guzmán-Barney, M. (2014). Natural co-infection of Solanum tuberosum crops by the Potato yellow vein virus and potyvirus in Colombia. Agron Colomb, 32(2), 213-223. http://dx.doi.org/10.15446/agron.colomb.v32n2.43968
Wang, B., Ma, Y., Zhang, Z., Wu, Z., Wu, Y., Wang, Q. & Li, M. (2011). Review. Potato viruses in China. Crop Prot, 30(9), 1117–1123. http://dx.doi.org/10.1016/j.cropro.2011.04.001
Cómo citar
APA
ACM
ACS
ABNT
Chicago
Harvard
IEEE
MLA
Turabian
Vancouver
Descargar cita
CrossRef Cited-by
1. Tanmaya Kumar Bhoi, Ipsita Samal, Prasanta Kumar Majhi, J. Komal, Deepak Kumar Mahanta, Asit Kumar Pradhan, Varun Saini, M. Nikhil Raj, Mohammad Abbas Ahmad, Partha Pratim Behera, Mangali Ashwini. (2022). Insight into aphid mediated Potato Virus Y transmission: A molecular to bioinformatics prospective. Frontiers in Microbiology, 13 https://doi.org/10.3389/fmicb.2022.1001454.
2. Andrea García, Mónica Higuita, Rodrigo Hoyos, Yuliana Gallo, Mauricio Marín, Pablo Gutiérrez. (2023). Prevalence of RNA viruses in certified, and informal potato seed tubers in the province of Antioquia (Colombia). Archives of Phytopathology and Plant Protection, 56(1), p.29. https://doi.org/10.1080/03235408.2023.2170208.
3. Magdalena CARA, Amani BEN SLIMEN, Enea MITRI, Orges CARA, Dajana FRASHERI, Jordan MERKURI, Giuseppe PARRELLA, Toufic ELBEAINO. (2025). Molecular Detection and characterization of viruses infecting greenhouse-grown tomatoes in Albania. Phytopathologia Mediterranea, 64(1), p.77. https://doi.org/10.36253/phyto-15811.
4. Zineb Belabess, Abdessalem Tahiri, Rachid Lahlali. (2025). From Symptoms to Solutions: A Deep Dive Into Potato Leaf Roll Virus Pathology. Journal of Crop Health, 77(1) https://doi.org/10.1007/s10343-024-01096-3.
5. Pablo Gutiérrez, Ary Rivillas, Daniel Tejada, Susana Giraldo, Andrea Restrepo, María Ospina, Susana Cadavid, Yuliana Gallo, Mauricio Marín. (2021). PVDP: A portable open source pipeline for detection of plant viruses in RNAseq data. A case study on potato viruses in Antioquia (Colombia). Physiological and Molecular Plant Pathology, 113, p.101604. https://doi.org/10.1016/j.pmpp.2021.101604.
6. KU REHMAN, MN KHALID, MS NAWAZ. (2020). PREVALENCE OF POTATO LEAF ROLL VIRUS DISEASE IMPACTS AND SEVERAL MANAGEMENT STRATEGIES TO HALT THE DAMAGE. Bulletin of Biological and Allied Sciences Research, 2020(1), p.21. https://doi.org/10.54112/bbasr.v2020i1.21.
7. Yuliana Marcela Gallo García, Andrea Sierra Mejía, Livia Donaire Segarra, Miguel Aranda, Pablo Andres Gutiérrez Sánchez, Mauricio Marín Montoya. (2019). Coinfección natural de virus de ARN en cultivos de papa (Solanum tuberosum subsp. Andigena) en Antioquia (Colombia). Acta Biológica Colombiana, 24(3), p.546. https://doi.org/10.15446/abc.v24n3.79277.
8. Andrea Sierra, Yuliana Gallo, Meike Estrada, Pablo Gutiérrez, Mauricio Marín. (2021). Detection of four RNA viruses in commercial and informal potato seed tubers in Antioquia (Colombia). Archives of Phytopathology and Plant Protection, 54(5-6), p.273. https://doi.org/10.1080/03235408.2020.1829424.
9. Yuliana Gallo, Mauricio Marín, Pablo Gutiérrez. (2021). Detection of RNA viruses in Solanum quitoense by high-throughput sequencing (HTS) using total and double stranded RNA inputs. Physiological and Molecular Plant Pathology, 113, p.101570. https://doi.org/10.1016/j.pmpp.2020.101570.
10. Yuliana Marcela Gallo García, Andrea Sierra Mejia, Pablo Andrés Gutiérrez Sánchez, Mauricio Alejandro Marín Montoya. (2020). Prevalencia de cinco virus de ARN en tubérculos-semilla de papa cultivados en Antioquia (Colombia). Biotecnología en el Sector Agropecuario y Agroindustrial, 19(1), p.66. https://doi.org/10.18684/bsaa.v19.n1.2021.1343.
Dimensions
PlumX
Visitas a la página del resumen del artículo
Descargas
Licencia
Derechos de autor 2017 Acta Agronómica

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Política sobre Derechos de autor:Los autores que publican en la revista se acogen al código de licencia creative commons 4.0 de atribución, no comercial, sin derivados.
Es decir, que aún siendo la Revista Acta Agronómica de acceso libre, los usuarios pueden descargar la información contenida en ella, pero deben darle atribución o reconocimiento de propiedad intelectual, deben usarlo tal como está, sin derivación alguna y no debe ser usado con fines comerciales.