Publicado
Modelling of 3D-Structures of the Rare Melanocortin-1-Receptor Mutations Associated to Melanism in the Bananaquit
Modelado de estructuras-3D de mutaciones raras del receptor-1-melanocortina asociadas al melanismo en el mielero
MODELAGEM DE ESTRUTURAS-3D DE MUTAÇÕES RARAS DO RECEPTOR-1-MELANOCORTINA ASSOCIADAS AO MELANISMO NO MIELERO
DOI:
https://doi.org/10.15446/abc.v26n1.81432Palabras clave:
Birds, Eumelanin, E92K, Protein Evolution, 3D-folding (en)Aves, Eumelanina, Evolución de Proteínas, E92K, Torción-3D (es)
Pássaros, Eumelanina, Evolução de Proteínas, E92K, Torción-3D (pt)
Descargas
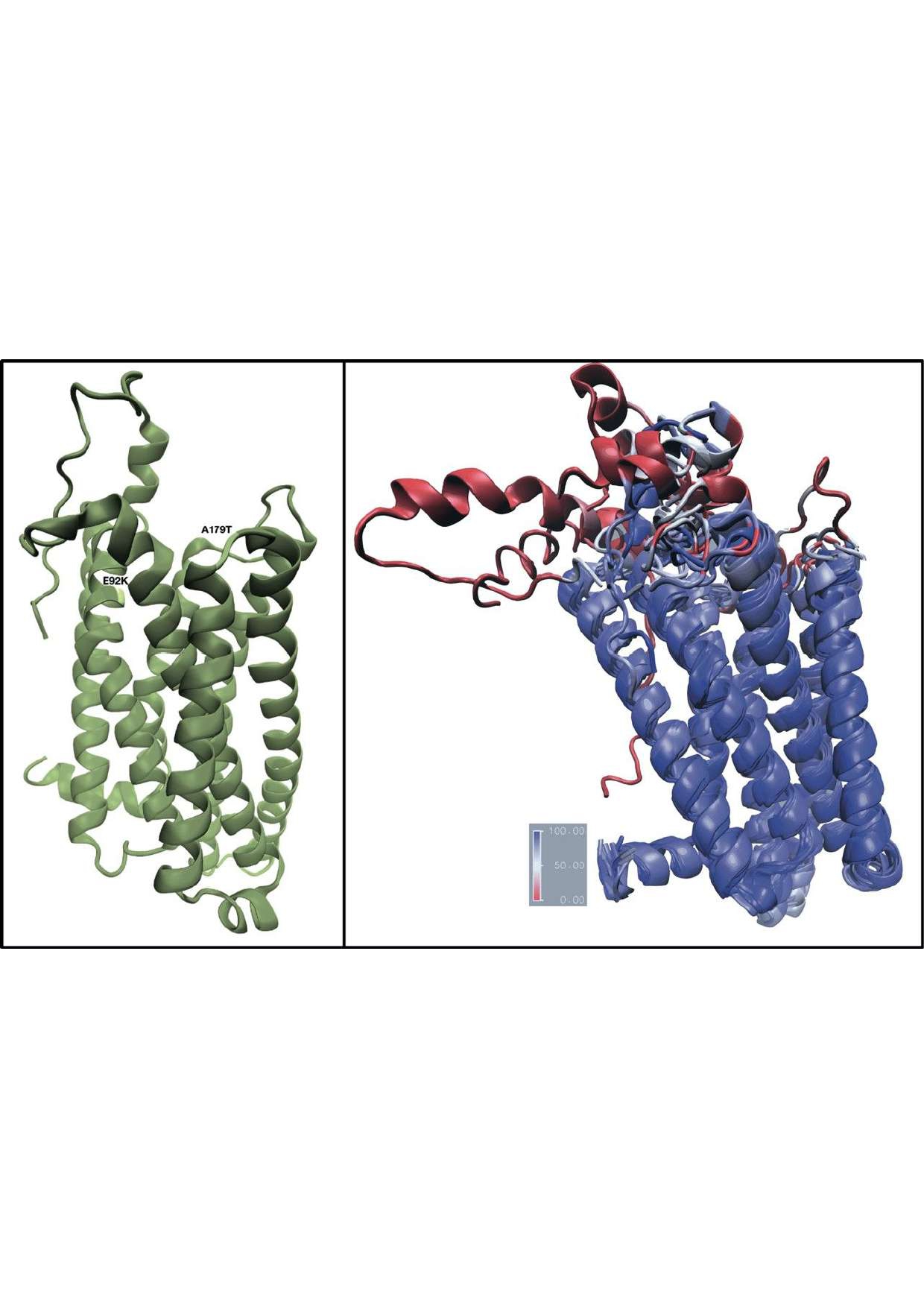
Melanism in plumage color is often associated to the single nucleotide polymorphism of the melanocortin-1-receptor (MC1R). Despite the striking association between the substitution of a Glutamic-acid by for a Lysine at position 92 on the MC1R protein and a completely black plumage, an in-depth understanding of the effect of missense mutations on the conformational change and behavior of the MC1R in the lipid bilayer caused by the absence of a crystal structure is lacking. We examine the structural basis for receptor activation using DNA sequences from the GenBank to perform in silicoprotein homology-based modeling. Our tridimensional model shows that the Alanine for a 179-Threoninesubstitution is a structural complement of the charge-reversing effect associated to the substitution of a Glutamic-acid by for a Lysine at position 92 on the MC1R. We proposed the possibility of gradual evolution in stability and electrostatic properties of the MC1R by the sequential accumulation of these two rare substitutions. These two rare substitutions further perturb physical-chemical properties that may be necessary folding requirements of the constitutively active MC1R forms without altering of ligand binding affinity. The computational coarse-grained molecular dynamics of the MC1R binding affinities to the melanocyte-stimulating hormone predicted the disparity in ligand binding amongalleles. We speculate that the disparity in structural constraints and ligand binding among the alleles within heterozygous individuals may contribute as a mechanism to the plumage color variation in the Coereba flaveola.
El melanismo en el color del plumaje se asocia frecuentemente al polimorfismo del receptor melanocortina-1(MC1R). La ausencia de una estructura cristalográfica de la asociación entre la sustitución del Glutamato por Lisina en la posición 92 de la proteína MC1R y el plumaje completamente negro, no ha permitido tener un mejor entendimiento del efecto de mutaciones no sinónimas en la conformación y en el comportamiento en la membrana del MC1R. Examinamos la estructura asociada a la activación del receptor usando secuencias de ADN obtenidas del GenBank, para un modelamiento in silicode formas homólogas de la proteína. El modelo tridimensional muestra que la sustitución de Alanina por la Treonina en la posición 179 es un complemento estructural al efecto de reversión de carga asociado a la sustitución del Glutamato por Lisina en la posición 92 del MC1R. Proponemos la posibilidad de evolución gradual de la estabilidad y de propiedades electrostáticas del MC1R por la acumulación de estas substituciones. Estas perturban las propiedades fisicoquímicas que podrían ser necesarias para el plegamiento de las formas constitutivamente activas del MC1R sin alterar la afinidad de empalme con el ligando. La modelación computacional de la dinámica molecular de la afinidad de empalme del MC1R a la hormona estimulante de meloncitos predice la disparidad de la unión con el ligando entre alelos. Consideramos que posiblemente la disparidad entre alelos en heterocigotos en cuanto a restricciones estructurales y la unión con el ligando podría contribuir a la variación en el color del plumaje en Coereba flaveola.
Referencias
Abolins-Abols M, Kornobis E, Ribeca P, Wakamatsu K, Peterson MP, Ketterson ED, et al., A role for differential gene regulation in the rapid diversification of melanic plumage coloration in the dark-eyed junco (Junco hyemalis). BioRxiv [Internet]. 2018(May):315762. Doi: https://doi.org/10.1101/315762
Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. Doi: https://doi.org/10.1038/nature02263
Beaumont KA, Newton RA, Smit DJ, Leonard JH, Stow JL, Sturm RA. Altered cell surface expression of human MC1R variant receptor alleles associated with red hair and skin cancer risk. Hum Mol Genet. 2005;14(15):2145–2154. Doi: https://doi.org/10.1093/hmg/ddi219
Beaumont KA, Shekar SL, Newton RA, James MR, Stow JL, Duffy DL, et al., Receptor function, dominant negative activity and phenotype correlations for MC1R variant alleles. Hum Mol Genet. 2007;16(18):2249–2260. Doi: https://doi.org/10.1093/hmg/ddm177
Bellemain E, Gaggiotti OE, Fahey A, Bermingham E, Ricklefs RE. Demographic history and genetic diversity in West Indian Coereba flaveola populations. Genetica. 2012;140(4–6):137–148. Doi: https://doi.org/10.1007/s10709-012-9665-6
Benkert P, Tosatto SCE, Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins Struct Funct Genet. 2008;71(1):261–277. Doi: https://doi.org/10.1002/prot.21715
Benned-Jensen T, Mokrosinski J, Rosenkilde MM. The E92K melanocortin 1 receptor mutant induces camp production and arrestin recruitment but not ERK activity indicating biased constitutive signaling. PLoS One. 2011;6(9):e24644. Doi: https://doi.org/10.1371/journal.pone.0024644
Berezovsky IN, Tumanyan VG, Esipova NG. Representation of amino acid sequences in terms of interaction energy in protein globules. FEBS Lett. 1997;418(1-2):43–46. Doi: https://doi.org/10.1016/S0014-5793(97)01346-X
Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, et al., The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. Doi: https://doi.org/10.1093/nar/28.1.235
Charif D, Lobry JR. SeqinR 1.0-2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis. 2007 p. 1–26. Doi: https://doi.org/10.1007/978-3-540-35306-5_10
Cherezov V, Hanson MA, Stevens RC, Rosenbaum DM, Rasmussen SGF, Foon ST, et al., High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. Doi: https://doi.org/10.1126/science.1150577
Cibois A, Thibault JC, Pasquet E. The molecular basis of the plumage colour polymorphism in the Tahiti reed-warbler Acrocephalus caffer. J Avian Biol. 2012;43(1):3–8. Doi: https://doi.org/10.1111/j.1600-048X.2011.05546.x
Darden T, York D, Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. Doi:https://doi.org/10.1063/1.464397
Guruprasad K, Reddy B, Pandit M. Correlation between the stability of a protein and its dipeptide composition : A novel approach for predicting in vivo stability from its primary structure. Protein Eng. 1990;4(2):155–161. Doi: https://doi.org/10.1093/protein/4.2.155
Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity (Edinb). 2006;97:222–234. Doi: https://doi.org/10.1038/sj.hdy.6800861
Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A Single Amino Acid Mutation Contributes to Adaptive Beach Mouse Color Pattern. Science. 2006;313(5783):101–104. Doi: https://doi.org/10.1126/science.1126121
Holder JR, Haskell-Luevano C. Melanocortin Ligands: 30 Years of Structure-Activity Relationship (SAR) Studies. Med Res Rev. 2004;24(3):325–356. Doi: https://doi.org/10.1002/med.10064
Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268(5214):1144–1149. Doi: https://doi.org/10.1126/science.7761829
Humphrey W, Dalke A, Schulten K. VMD: Visual Molecular Dynamics. J Mol Graph. 1996;14(1):33–38. Doi: https://doi.org/10.1016/0263-7855(96)00018-5
Ibarrola-Villava M, Peña-Chilet M, Llorca-Cardeñosa MJ, Oltra S, Cadenas CM, Bravo J, et al., Modeling MC1R rare variants: A structural evaluation of variants detected in a mediterranean case-control study. J Invest Dermatol. 2014;134(4):1146–1149. Doi: https://doi.org/10.1038/jid.2013.469
Jackson IJ. Homologous pigmentation mutations in human, mouse and other model organisms. Hum Mol Genet. 1997;6(10):1613–1624. Doi: https://doi.org/10.1093/hmg/6.10.1613
Jacobs GH. Losses of functional opsin genes, short-wavelength cone photopigments, and color vision - A significant trend in the evolution of mammalian vision. Vis Neurosci. 2012;30(1–2):39–53. Doi: https://doi.org/10.1017/S0952523812000429
Janin J, Sternberg MJE. Protein flexibility, not disorder, is intrinsic to molecular recognition. F1000 Biol Rep. 2013;5(2):1–7. Doi: https://doi.org/10.3410/b5-2
Kerje S, Lind J, Schutz K, Jensen P, Andersson L. Melanocortin 1-receptor (MC1R) mutations are associated with plumage colour in chicken. Anim Genet. 2003;34(4):241–248. Doi: https://doi.org/10.1046/j.1365-2052.2003.00991.x
Krishnan S, Cryberg RL. Effects of mutations in pigeon Mc1r implicate an expanded plumage color patterning regulatory network. BioRxiv. 2019:1–36. Doi: https://doi.org/10.1101/792945
Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. Doi: https://doi.org/10.1016/0022-2836(82)90515-0
Lennard-Jones JE, Strachan C. Material Forces and Configurational Forces in the Interaction of Elastic Singularities. Proc R Soc Lond A Math Phys Sci. 1935;150(870):442–455. Doi: https://doi.org/10.1098/rspa.1935.0114
Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J Mol Model. 2001;7(8):306–317. Doi: https://doi.org/10.1007/S008940100045
Ling MK, Lagerström MC, Fredriksson R, Okimoto R, Mundy NI, Takeuchi S, et al., Association of feather colour with constitutively active melanocortin 1 receptors in chicken. Eur J Biochem. 2003;270(7):1441–1449. Doi: https://doi.org/10.1046/j.1432-1033.2003.03506.x
Lu D, Vage DI, Cone RD. A Ligand-Mimetic Model for Constitutive Activation of the Melanocortin-1 Receptor. Mol Endocrinol. 1998;12(4):592–604. Doi: https://doi.org/10.1210/mend.12.4.0091
Matsumoto Y, Hiramatsu C, Matsushita Y, Ozawa N, Ashino R, Nakata M, et al., Evolutionary renovation of L/M opsin polymorphism confers a fruit discrimination advantage to ateline New World monkeys. Mol Ecol. 2014;23(7):1799–1812. Doi: https://doi.org/10.1210/10.1111/mec.12703
Moore DS. Amino acid and peptide net charges: A simple calculational procedure. Biochem Educ. 1985;13(1):10–11. Doi: https://doi.org/10.1016/0307-4412(85)90114-1
Mundy NI. A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc R Soc B Biol Sci. 2005;272(1573):1633–1640. Doi: https://doi.org/10.1098/rspb.2005.3107
Nadeau NJ, Minvielle F, Mundy NI. Association of a Glu92Lys substitution in MC1R with extended brown in Japanese quail (Coturnix japonica). Anim Genet. 2006;37(3):287–289. Doi: https://doi.org/10.1111/j.1365-2052.2006.01442.x
Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, et al., Crystal Structure of Rhodopsin: A G Protein-Coupled Receptor. Science. 2000;289(5480):739–745. Doi: https://doi.org/10.1126/science.289.5480.739
Paynter RA. Check-list of birds of the world. [Internet]. vol XIV. Cambridge: Cambridge :Harvard University Press; 1968. p. 1-433. Disponible en: https://www.biodiversitylibrary.org/page/14481194#page/7/mode/1up
Ponder JW, Case DA. Force Fields For Protein Simulations. Adv Protein Chem. 2003; 66: 27–85. Doi: https://doi.org/10.1016/S0065-3233(03)66002-X
Ran J-S, You X-Y, Jin J, Zhou Y-G, Wang Y, Lan D, et al., The Relationship between MC1R Mutation and Plumage Color Variation in Pigeons. Biomed Res Int. Hindawi Publishing Corporation. 2016;2016:2–7. Doi: https://doi.org/10.1155/2016/3059756
Rees JL. Review - Pigment Gene Focus The Melanocortin 1 Receptor ( MC1R ): More Than Just Red Hair. Cell Res. 2000;13:135–140. Doi: https://doi.org/10.1034/j.1600-0749.2000.130303.x
Ringholm A, Klovins J, Rudzish R, Phillips S, Rees JL, Schiöth HB. Pharmacological characterization of loss of function mutations of the human melanocortin 1 receptor that are associated with red hair. J Invest Dermatol. 2004;123(5):917–923. Doi: https://doi.org/10.1111/j.0022-202X.2004.23444.x
Robbins LS, Nadeau JH, Johnson KR, Kelly MA, Roselli-rehfuss L, Baack E, et al., Pigmentation Phenotypes of Variant Extension Locus Alleles Result from Point MutationsThat Alter MSH Receptor Function. Cell. 1993;72(6):827–834. Doi: https://doi.org/10.1016/0092-8674(93)90572-8
Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:1–11. Doi: https://doi.org/10.1186/1471-2105-7-382
Shahlaei M, Mousavi A. A 3D Model for human melanocortin 4 receptor refined with molecular dynamics simulation. J Reports Pharm Sci. 2014;3(1):42–53. Doi: https://doi.org/10.22110/jrps.v3i1.1677
Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33((Web Server issue):W244–W248. Doi: https://doi.org/10.1093/nar/gki408
Spiliotopoulos D, Spitaleri A, Musco G. Exploring PHD Fingers and H3K4me0 Interactions with Molecular Dynamics Simulations and Binding Free Energy Calculations: AIRE-PHD1, a Comparative Study. PLoS One. 2012;7(10):e46902. Doi: https://doi.org/10.1371/journal.pone.0046902
Suryaningsih BE, Sadewa AH, Wirohadidjojo YW, Soebono H. Association between heterozygote Val92Met MC1R gene polymorphisms with incidence of melasma: A study of Javanese women population in Yogyakarta. Clin Cosmet Investig Dermatol. 2019;12:489–495. Doi: https://doi.org/10.2147/CCID.S206115
Takeuchi S, Suzuki H, Yabuuchi M, Takahashi S. Possible involvement of melanocortin 1-receptor in regulating feather color pigmentation in the chicken. Biochim Biophys Acta - Gene Struct Expr. 1996;1308(2):164–168. Doi: https://doi.org/10.1016/0167-4781(96)00100-5
Tan K, Pogozheva ID, Yeo GSH, Hadaschik D, Keogh JM, Haskell-Leuvano C, et al., Functional characterization and structural modeling of obesity associated mutations in the melanocortin 4 receptor. Endocrinology. 2009;150(1):114–125. Doi: https://doi.org/10.1210/en.2008-0721
Teilum K, Olsen JG, Kragelund BB. Protein stability, flexibility and function. Biochim Biophys Acta - Proteins Proteomics. Elsevier B.V.; 2011;1814(8):969–976. Doi: https://doi.org/10.1016/j.bbapap.2010.11.005
Theron E, Hawkins K, Bermingham E, Ricklefs RE, Mundy NI. The molecular basis of an avian plumage polymorphism in the wild. Curr Biol. 2001;11(8):550–557. Doi: https://doi.org/10.1016/s0960-9822(01)00158-0
Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins Struct Funct Genet. 2000;41(3):415–427. Doi: https://doi.org/10.1002/1097-0134(20001115)41:3<415::AID-PROT130>3.0.CO;2-7
De Vries SJ, Van Dijk M, Bonvin AMJJ. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. Doi: https://doi.org/10.1038/nprot.2010.32
Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17(4):263–270. Doi: https://doi.org/10.1002/humu.22
Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(40):1–8. Doi: https://doi.org/10.1186/1471-2105-9-40
Cómo citar
APA
ACM
ACS
ABNT
Chicago
Harvard
IEEE
MLA
Turabian
Vancouver
Descargar cita
Licencia
Derechos de autor 2020 Acta Biológica Colombiana

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-CompartirIgual 4.0.
1. La aceptación de manuscritos por parte de la revista implicará, además de su edición electrónica de acceso abierto bajo licencia Attribution-NonCommercial-ShareAlike 4.0 (CC BY NC SA), la inclusión y difusión del texto completo a través del repositorio institucional de la Universidad Nacional de Colombia y en todas aquellas bases de datos especializadas que el editor considere adecuadas para su indización con miras a incrementar la visibilidad de la revista.
2. Acta Biológica Colombiana permite a los autores archivar, descargar y compartir, la versión final publicada, así como las versiones pre-print y post-print incluyendo un encabezado con la referencia bibliográfica del articulo publicado.
3. Los autores/as podrán adoptar otros acuerdos de licencia no exclusiva de distribución de la versión de la obra publicada (p. ej.: depositarla en un archivo telemático institucional o publicarla en un volumen monográfico) siempre que se indique la publicación inicial en esta revista.
4. Se permite y recomienda a los autores/as difundir su obra a través de Internet (p. ej.: en archivos institucionales, en su página web o en redes sociales cientificas como Academia, Researchgate; Mendelay) lo cual puede producir intercambios interesantes y aumentar las citas de la obra publicada. (Véase El efecto del acceso abierto).