Estimates of parameters, prediction and selection of an oil palm population in Ecuador
Estimativas de parámetros, predicción y selección en progenies de palma aceitera en Ecuador
DOI:
https://doi.org/10.15446/rfna.v71n2.71928Keywords:
Elaeis guineensis Jacq, Genetic parameters, Blup, Clusters, Selection index (en)Elaeis guineenses Jacq, Parámetros genéticos, Blup, Agrupamiento, Índice de selección (es)
Was used the REML/BLUP method to estimate the genetic parameters, select the best plants of Dura x Dura full-sib families, study the trait correlations, group families by multivariate similarity and to determine the number of repeated measurements required for the selection of the traits (bunch number, fresh fruit bunch yield and average bunch weight). Twenty-four families developed in three trials were tested, together with one control per test from the genebank of the experimental station of Santo Domingo - INIAP in Ecuador. The evaluation lasted five years and was arranged in a randomized block design with 12 plants per plot and four replications. The population variability for traits and genetic heritability between plots was close to that found within plots. The genetic gain of the 10 selected plants, was 43% higher than the overall average. The correlation was low and negative only between bunch number and average bunch weight. By Tocher cluster analysis, six groups were formed, and in group IV, families selected by average ranking (3A, 5C, and 7B). It was concluded that the BLUP estimates are encouraging with a view to a continuous breeding program of oil palm, with the possibility of maximizing genetic gains in future generations.
Recibido: 11 de octubre de 2016; Aceptado: 23 de noviembre de 2017
ABSTRACT
Was used the REML/BLUP method to estimate the genetic parameters, select the best plants of Dura x Dura full-sib families, study the trait correlations, group families by multivariate similarity and to determine the number of repeated measurements required for the selection of the traits (bunch number, fresh fruit bunch yield and average bunch weight). Twenty-four families developed in three trials were tested, together with one control per test from the genebank of the experimental station of Santo Domingo - INIAP in Ecuador. The evaluation lasted five years and was arranged in a randomized block design with 12 plants per plot and four replications. The population variability for traits and genetic heritability between plots was close to that found within plots. The genetic gain of the 10 selected plants, was 43% higher than the overall average. The correlation was low and negative only between bunch number and average bunch weight. By Tocher cluster analysis, six groups were formed, and in group IV, families selected by average ranking (3A, 5C and 7B). It was concluded that the BLUP estimates are encouraging with a view to a continuous breeding program of oil palm, with the possibility of maximizing genetic gains in future generations.
Keywords:
Elaeis guineensis Jacq, Genetic parameters, Blup, Clusters, Selection index.RESUMEN
Fue empleado el método REML/BLUP para estimar los parámetros genéticos, y seleccionar los mejores individuos provenientes de una población de hermanos germanos de Dura x Dura, a partir de un análisis de correlación entre caracteres, realizando un agrupamiento de familias por disimilaridad multivariada y determinación del número de medidas repetidas necesarias para la selección de las características (número y peso medio de racimos). Fueron evaluadas 24 familias procedentes de tres ensayos del banco de germoplasma de la estación experimental Santo Domingo del INIAP en Ecuador. La evaluación fue realizada en un periodo de cinco años, empleando un diseño en bloques al azar, con doce plantas por parcela y cuatro repeticiones. La variabilidad de la población en relación a las características evaluadas y heredabilidad de individuos dentro del bloque, similar a la encontrada dentro de familias en las parcelas. La ganancia genética de las 10 plantas seleccionadas representa un 43% superior a la media general. La correlación fue baja y negativa para número de racimos y peso medio de racimos. Con base en el agrupamiento de Tocher se obtuvieron seis grupos, donde el grupo IV agrupa las familias seleccionadas por el Rank-medio multivariado (3A, 5C y 7B). Se puede concluir que las estimativas obtenidas por el BLUP, estimulan la continuidad del programa de mejoramiento genético de racimos, con posibilidad de maximizar las ganancias genéticas en generaciones futuras.
Palabras clave:
Elaeis guineenses Jacq, Parámetros genéticos, Blup, Agrupamiento, Índice de selección.The oil palm (Elaeis guineensis Jacq.), known as oil palm, "palma-aceitera" (in Spanish) and "palmier à huile" (in French), belongs to the order Palmales and the family Arecaceae (ex-Palmae). It is grown mainly in Asia, Africa and Central and South America (Gomes et al., 2009). In 2010, the area with oil palm plantations in Ecuador was 248,199 hectares, of which 193,502 ha are actually producing. The main palm-oil-producing provinces are Esmeraldas (152,679 ha); Os Ríos (31,276 ha); Sucumbíos and Orellana (24,102 ha); Pichincha (16,363 ha); Santo Domingo (16,364 ha), Guayas (4195 ha); Cotopaxi (1280 ha); Manabí (1237 ha); Bolívar (155 ha) and Imbabura (23 ha) (INEC, 2010).
The world oil palm production is grown mostly on plantations in Malaysia and Indonesia (86%), and 5% are produced in countries of South America (Colombia, Ecuador and Brazil). According to FEDAPAL (2011), Ecuador produced 440,000 t of palm oil in 2011, of which 210,000 was destined for domestic consumption and 230,000 for export. In 2015, the global demand for vegetable oil is expected to be 170 million and the demand for palm oil 68 million tons. During the last years there have been few attempts to produce biodiesel by different methods from waste cooking oil (Meng et al., 2008), waste frying palm oil (Lertsathapornsuk et al., 2008), canola oil (Leung and Guo, 2006) soybean oil and sunflower seed oil (Yang and Xie, 2007).
The plant types of oil palm can be classified in three groups, according to the endocarp thickness: Dura, with a thick endocarp; Pisífera, fruits without endocarp and generally, abortive, and Tenera fruits with thin endocarp and greater proportion of mesocarp of the fruit than Dura (Ferreira et al., 2012).
In the palm breeding programs, two types of parents are used: Dura as female and Pisifera as male parent, to breed the hybrid Tenera. The Dura Deli population was derived from only four palms of the Botanical Garden in Bogor, Indonesia (Rosenquist, 1985). The existence of genetic variability in breeding programs of palm oil has been reported by several authors (Musa et al., 2004; Rafii et al., 2002). Okoye (2007) reported that high heritability estimate is correlated to the high genetic gain in bunch production, which is one of the most important characteristic for oil production, as well as the oil content in the bunch.
According to Cruz and Carneiro (2006), the genetic improvement depends on the correct choice of the best plants as parents of the following generations. The breeding of this crop is limited due to the long period of evaluation and the cost of this evaluation in order to achieve a cultivar. A selection cycle that includes evaluation and phenotypic selection, and crossing between the families selected to form a new population requires about 19 years (Wong and Bernardo, 2008). The selection of superior plants is based on several traits, which can be genetically and phenotypically correlated. Selection bias can occur if these traits are analyzed individually. In tests conducted in the germplasm bank of the experimental station Santo Domingo of INIAP to identify the best Dura mother plants, selection between and within families and combined selection were used (Ortega et al., 2008a).
To estimate the genetic parameters in progeny tests, variance was analyzed (ANOVA) and mean squares partitioned based on their mathematical expectations (Cruz and Carneiro, 2006), and on REML/BLUP (restricted maximum likelihood/best linear unbiased prediction). The BLUP procedure uses information of relatives and maximizes selective accuracy, providing greater genetic gains. The REML procedure is a valuable estimation of variance components, better than the method of analysis of variance (ANOVA) in several situations, particularly with unbalanced data (Resende, 2002).
Resende (2002) stated that the most appropriate procedure of prediction of breeding values in the genetic evaluation of perennials is the individual BLUP approach, consisting basically of the prediction of breeding values of the random effects of the statistical model associated to the phenotypic data, adjusting the data to the fixed effects and to the unequal number of data in the plots by the mixed model methodology (Henderson and Quaas, 1976).
Based on the above, this study aimed to optimize the oil palm breeding program in Ecuador by the estimation of genetic parameters and selection of the best plants from Dura x Dura full-sib families, by the study of trait correlations, the grouping of families by multivariate similarity and by determining the number of repeated measurements required for the selection of the traits number, bunch yield and average bunch weight, using the REML/BLUP method.
MATERIALS AND METHODS
For this study, we used data from a five-year assessment of full-sib families of oil palm (Elaeis guineensis Jacq), from the Germplasm Bank of the Experimental Station Santo Domingo of INIAP (Instituto Nacional de Investigaciones Agropecuarias), in Ecuador (km 38, via Santo Domingo-Quinindé; longitude 79°20'W, latitude 00°06'N; altitude 300 m).
The experiment was arranged in a randomized block design, with 12 plants per plot, 4 replications and triangular spacing of 9 m x 9 m. Twenty-four families derived from three trials were tested in the plots 3A (12 families), 13B (9 families) and Caseta (6 families) of Dura x Dura full-sibs, with one control per test. The traits bunch number (BNo), fresh fruit bunch yield (FFBy) and average bunch yield (ABWt) were evaluated.
The genotypic values of each trait were calculated by REML (Restricted Maximum Likelihood) applied to the linear mixed model and prediction of breeding values by BLUP (Best linear unbiased prediction) and, for the evaluation of 27 families (including controls), we used the computer program SELEGEN-REML/BLUP, Resende (2007). The statistical model: y= Xβ+Za+Wc+Tp+e was applied, where y is the data vector; β is the vector of fixed effects (means of blocks, tests and years) added to the overall average; a is the vector of additive genetic effects (assumed as random); c is the vector of the plot effects (assumed as random); p is the vector of permanent plant effects (assumed as random); and e is the error or residual vector (random). The letters X, Z, W and T are incidence matrices for β, a, c, and p, respectively. The mean and variance distributions and structures are:


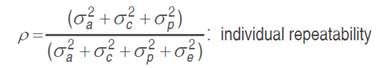
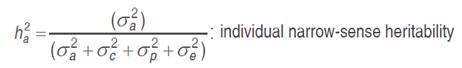
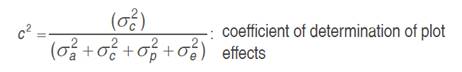

In view of the imbalance in the experiment, the deviance analysis (ANADEV) was performed using the likelihood ratio test (LRT). The deviances of the full model and reduced model were calculated, using the model with and without h2, c2 and p2 values for each variable, by subtracting the deviance of the reduced models from those of the full model, the likelihood ratio (LRT) tested by chi-square, at one degree of freedom and 1 and 5% probability.
Individual genetic parameters were calculated by REML: Vg: genotypic variance between full-sib progenies, (1/2) additive genetic variance, plus (1/4) of the dominance genetic variance, ignoring epistasis; Vplot: environmental variance among plots; Vperm: variance of permanent effects; Vwithin: residual variance within plot; Vph: individual phenotypic variance; h2a: individual narrow-sense heritability, ignoring the fraction (1/4) of the dominance genetic variance; c2plot: coefficient of determination of plot effects; c2perm: coefficient of determination of permanent effects; h2gd: genetic heritability within plot, ignoring the fraction of the dominance genetic variance; CVe: residual coefficient of variation; CVg: coefficient of genotypic variation among progenies; CVr coefficient of relative variation (CVg/CVe).
For the selection of the best families, the variable fresh fruit bunch yield (FFBy) was considered. Later, to increase the selection efficiency, the simultaneous selection of the three variables under study was taken into account, for which the selection index based on the sum of ranks (Mulamba and Mock, 1978) was used. For this purpose, the 27 families including three controls were ranked by the average rank index, combining all predicted breeding values of each trait.
The genotypic correlations between the studied traits were also analyzed. Cluster analysis was performed by the Tocher method described in detail by Cruz and Regazzi (1994). The mean Euclidean genetic distance was used as similarity measure between phenotypes. In addition, repeatability, efficiency of measurements of harvests in the five years of evaluation and the accuracy values were determined for each trait.
RESULTS AND DISCUSSION
Deviance analysis (ANADEV) was performed, considering the model adjusted for genotype, plot and permanent effect, which proved significant by the likelihood ratio test (LTR), at 1% and 5% probability by the chi-square test, for the traits bunch number (BNo), fresh fruit bunch yield (FFBy) and average bunch weight (ABWt) of oil palm (Table 1). The significant genotype effect indicated the existence of genetic variability among the 27 families in the three tests for the three traits under study. Regarding the plot effect, significant environmental differences were also demonstrated between plots in each replication and test. This analysis revealed the existence of highly significant permanent differences (permanent environmental and non-additive effects) among plants in the five years of evaluation.
Table 1: Deviance analysis (ANADEV) for the variables: bunch number, bunch yield and average bunch weight, in three tests of Dura x Dura full-sib progenies.
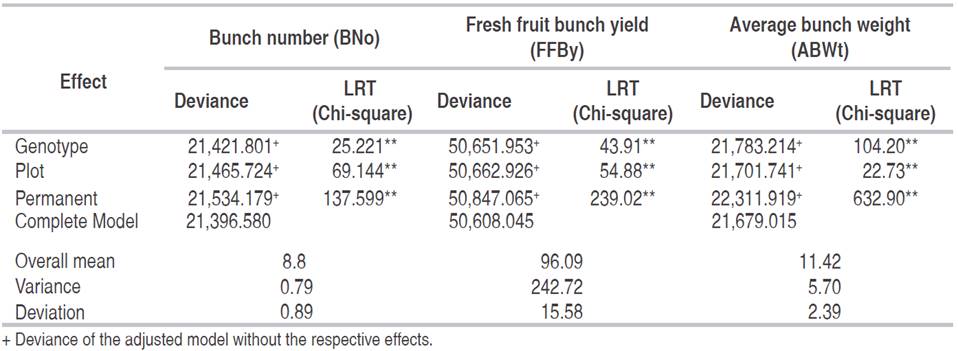
The estimated overall average with the respective deviation in the five years of evaluation was 8.8 ± 0.89 for trait BNo; 96.09 ± 15.58 kg for FFBy; and 11.42 ± 2.39 kg for ABWt (Table 1). The variance was 0.79, 5.70 and 242.7 for BNo, ABWt and FFBy, respectively.
The estimated genetic parameters and coefficients of determination of the studied variables are shown in Table 2. For genotypic variance, only the additive fraction was considered, estimated at 1.11 for BNo, 309 for FFBy and 6.52 for ABWt, corresponding to a total variance of 7%, 11% and 27%, respectively (Table 2). Lopes et al. (2012) in hybrids caiaué with oil palm was estimated genotypic variance 3.24 BNo, 554.28 FFBy and 5.92 ABWt. For plot variance, the estimates were less than 5% of the total variance for the three traits under study, indicating efficient control in the plots. However, the permanent variance was estimated at 1.91 for BNo and 436.66 for FFBy, each corresponding to less than 15% of the total variance. For ABWt, permanent variance was 5.08, representing 21% of the total variability. In general, differences in plants between years of evaluation in harvests were stated, mainly due to the strong environmental influence. This result is consistent with Corley and Gray (1976b), who pointed out that in the first years bunches weigh on average 5 kg, but reach 25 kg after 15 years, whereas the BNo tends to decreases over the years (Lim and Chan, 1998).
Table 2: Estimates of genetic parameters for the variables: bunch number, bunch yield and average bunch weight in three trials of Dura x Dura full-sib progenies.

The values of the within-plot variance were 12.67 for BNo 2004.9 for FFBy and 11.82 for ABWt, representing 76%, 69% and 49% of the total variance, respectively. This indicated that the greatest variability is within plots, due to environmental effects within each plot distributed in the three tests (distance between plants). The narrow-sense or additive heritability of 13.3% for the variable BNo (Table 2) (versus 21% for FFBy and 54% for ABWt) was considered relatively low. Based on these estimates, to ensure the selection of the best plants and increased genetic gain in breeding programs, it is recommended that the traits FFBy and ABWt should be primarily taken into account. However, it should be mentioned that the genetic variability in the population is low, due to the narrow genetic base of oil palm. Corley and Tinker (2009) and Kwong et al. (2016), reported the loss of genetic variation in Dura Deli oil palm populations after generations of selection with limited genetic base. By BLUP, Soh (1994) determined values of individual heritability in the narrow sense of 35% for BNo and 20% for FFBy.
It was also observed that the genetic heritability within plots was lower for BNo and FFBy (8.8% and 15%, respectively), with ABWt ratio was 55% similar value found in the additive heritability. This demonstrates that for these two variables, variance within plots was influenced by the environment, hampering the selection of the best plants. In the case of ABWt, it was found that the trait is related to BNo and FFBy. Sparnaaij (1969), Ooi et al. (1973) and Van der Vossen, (1974) reported that BNo is negatively correlated with ABWt. The use of additive heritability would therefore be more convenient to increase the selection efficiency of plants among than within the best families. Selection between and within families takes into consideration the best family, and later, it selects the best individual within the selected family (Bhering et al., 2013).
The coefficient of determination of the plot effects (c²plot) (Table 2) was low (0.06 for BNo, 0.05 for FFBy and 0.03 for ABWt), revealing that the experimental design was adequate and that homogeneity within blocks was achieved. The permanent plot effect reflects variations in the environment between plots within blocks. Changes related to the environment are caused by circumstances which affect the plant permanently. It was found that the individual permanent effect (c²perm) was little variable (0.11, 0.15 and 0.21), with a low magnitude for BNo and a weak permanent influence of the environment on the phenotypic expression of those traits.
The experimental coefficient of variation (CVe%) was low (10% - 21%), demonstrating good accuracy and a low error rate of the experimental measurements. Okoye et al. (2009) the experimental coefficient of variation fluctuate 16.1% - 37.8%, in material Dura x Tenera. The coefficients of genetic variation among families were higher (12% - 22%) than the experimental coefficient of variation, demonstrating the existence of predominantly genetic variability in the studied traits. The relative variation coefficient (CVr%) for the three variables was higher than 1. According to Resende and Duarte (2007) and Noh et al. (2014), with four blocks or replications, the accuracy was 89%, demonstrating that the experiment was appropriate, allowing greater efficiency when selecting the best families, in agreement with the additive heritability estimated between families, which were also high in relation to the heritability within families (Table 2).
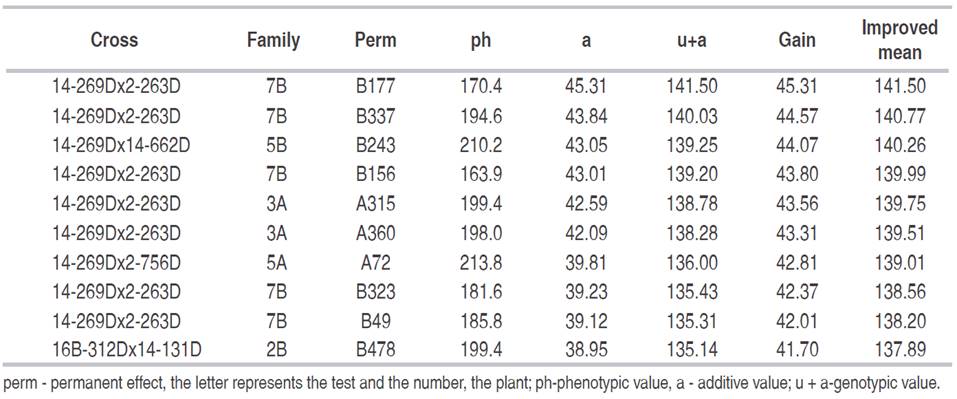
The best 10 plants were selected based on the trait fresh fruit bunch yield (Table 3), of the seven plants from test 13B, five were from family 7B, one of family 5B and one of family 2B, while of the three plants of test 3A, two belong to family 3A and one to family 5A. It is noteworthy that the five plants of family 7B and two of family 3A were from the same cross (14-269D x 2-263D) and that the plants of family 5B (14-269Dx14-662D) and 5A (14-269Dx2-756D) also have parent 14-269D in common. The average phenotypic values of selected plants ranged from 163.9 to 213.8 kg. The estimated additive effects ranged from 38.95 to 45.31 kg and the additive genetic values from 135.1 to 141.5 kg. The genetic gain in the 10 plants for the variable yield was 41.70 kg that added of the overall average of 96.09 kg (Table 1), the improved population mean for the following generation will be 137.89 kg. Therefore, the genetic gain will be 43.4%. Using selection among and within full-sib families, Ortega et al. (2008a) estimated gains of 4.42% and of 4.92% by combined selection for the trait FFBy, considering only one of the tests evaluated in this study.
Table 3: Selection of the 10 best plants for potential parents and crosses for the trait bunch yield, additive genetic effect, additive genetic values, selection gain and improved mean, in three tests of Dura x Dura full-sib progenies.
The genetic correlation coefficients between the traits bunch number, bunch yield and average bunch weight were estimated (Table 4). In general, the correlation estimates were low, except for FFBy and ABWt (0.78). However, the association between BNo and ABWt was low (-0.37), indicating greater variability in weight than bunch number. According to Ooi et al. (1973), Van der Vossen (1974), Cros et al. (2015), Kwong et al. (2017). BNo and ABWt are negatively correlated, since carbohydrate supply is limiting to the photosynthetic yield, so that a higher BNo after selection will inevitably reduce the ABWt, due to a compensation effect. It should be remembered that at the beginning of oil palm production, the BNo is high, but the ABWt is low. As the plants develop, the BNo decreases and FFBy increases. In Dura oil palm plants evaluated from 1992 to 1996, Ortega et al. (2008b) estimated negative genetic correlations between BNo and FFBy, except in 1994, when the correlation was positive and similar (0.21) to the estimated (0.27). These results indicate a strong influence of the environment on the expression of these traits, so that family selection to improve the population Dura should be based on the three traits simultaneously.
Table 4: Estimation of genetic correlations between bunch number, bunch yield and average bunch weight, based on three tests of Dura x Dura full-sib progenies.

To select families for the improvement of the population Dura, the 27 full-sib families (including the three controls) were sorted by the average rank based on estimated breeding values for each of the three study variables (Table 5). However, according to Ramalho and Lambert (2004), Kwong et al. (2017) the success of intrapopulation improvement depends on the crossing of plants of the best but most divergent families. Thus, the 27 full-sib families were grouped by dissimilarity (Table 6), based on the standardized mean Euclidean distance. Six groups were differentiated, of which group I contained most (15) families. Group IV comprised the best three families (3A, 5C and 7B), with the lowest average ranks (Table 5).
Table 5: Ranking of families based on the breeding value, Mulamba-rank selection index and rank-average for the traits bunch number, fresh fruit bunch yield and average bunch weight, in three tests of Dura x Dura full-sib progenies.
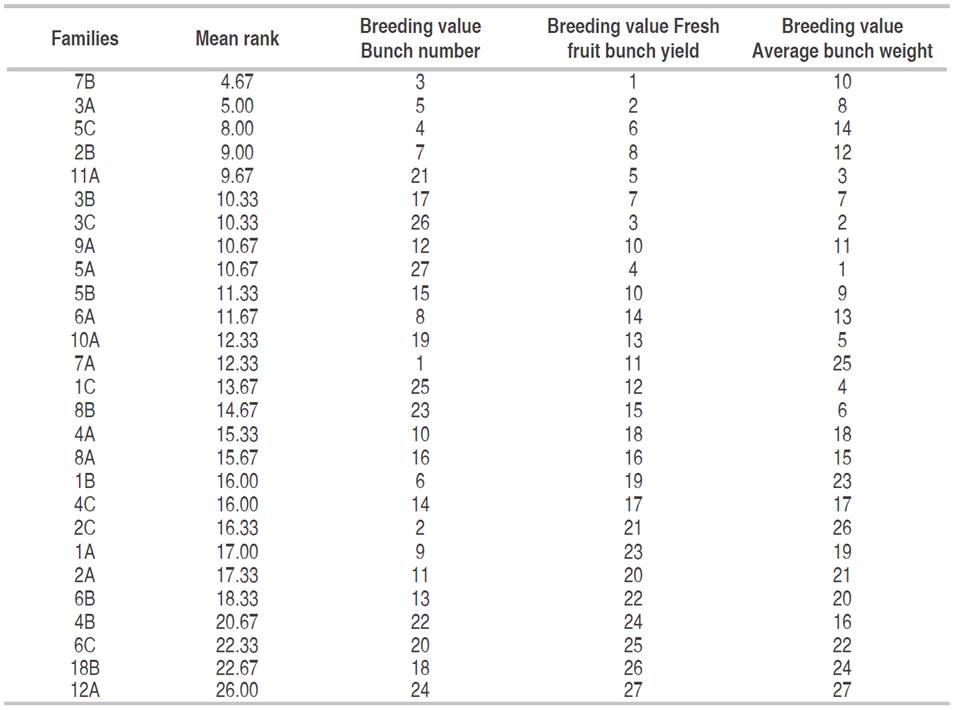
Table 6: Clustering of families assessed by the Tocher method, using the mean Euclidean genetic distance.

It was observed that the families 6C and 18B, used as controls in test 13B, and Caseta were allocated in group III. These two families were derived from the cross 1050D x 14-13A-491P and the third control (cross 14-93D x 13A-527P), evaluated in test 3A, was allocated in group VI. Thus, the selection of plants for recombination initially identified the best-performing families for the traits BNo, FFBy and ABWt simultaneously (Table 5), in each similarity group (Table 6), by the average rank values.
Subsequently, the plants were selected from these families with an estimated breeding value greater than 34 (Table 7). Family 7A of similarity group V, although better performing than the 2C family, was not included among the plants selected for recombination since the breeding values of the two best plants were lower than 21, occupying positions 108 and 141, respectively, in the ranking of breeding values. To determine the minimum number of harvests required to assess oil palm plants, the coefficients of repeatability, the coefficient of determination, the permanent accuracy and efficiency of evaluation for a single measurement were estimated for the three studied variables (Table 8). For the trait BNo, the repeatability for a single harvest was 0.31, with an accuracy of 0.27 and a correlation coefficient of 0.08, which is considered low. However, after five harvests, repeatability increased to 0.69, accuracy to 0.54 and the coefficient of determination to 0.29, with an efficiency of 96%.
Table 7: Selection of the 10 best plants for a new recombination cycle based on mean rank of the additive breeding value of the trait bunch yield (mean kg) and group similarity.
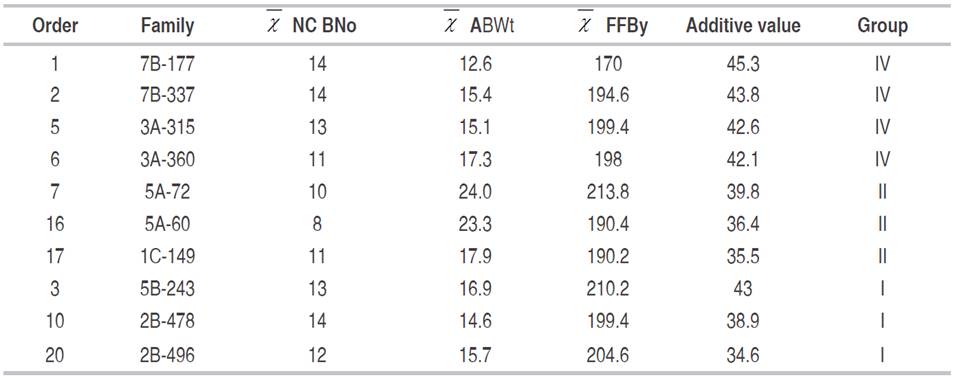
Table 8: Number of harvests needed for the variables bunch number, fresh fruit bunch yield and average bunch weight to achieve certain accuracy values.
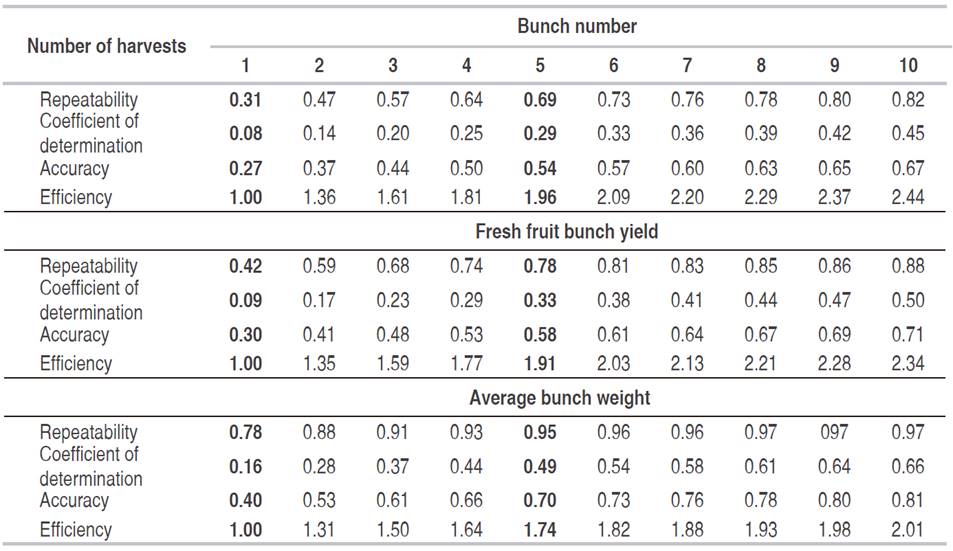
For FFBy, repeatability was 0.42, accuracy 0.30 and the coefficient of determination 0.09 for a single harvest. These values were higher than those for BNo. After five harvests however, repeatability was 0.78, accuracy 0.58 and the coefficient of determination 0.33, with an efficiency of 91% (Table 8). According to estimations of Ortega et al. (2008b), for FFBy in Dura progenies, the repeatability coefficient was 0.64 - 0.74, with a correlation coefficient of 0.90 - 0.94, by the methods ANOVA and principal components.
With regard to ABWt, the repeatability, accuracy and determination coefficients were higher (0.78, 0.40 and 0.16, respectively) than for the other variables, assuming a single harvest. In the case of five harvests, the repeatability, accuracy and coefficient of determination increased to 0.95, 0.70 and 0.49, respectively. However, the efficiency of assessment for five harvests decreased to 74%. Thus, to evaluate full-sib progenies of oil palm, it is recommended to evaluate the bunch yield and bunch number in five harvests.
CONCLUSIONS
The detected genetic variability and heritability between plots encourage continuous breeding programs of oil palm, with the possibility of maximizing gains in future generations.
The selection of plants based on the variable fresh fruit bunch yield-FFBy should be prioritized for satisfactory gains.
The correlations for bunch number-BNo, with fresh fruit bunch yield -FFBy and with - average bunch weight-ABWt were low, while for fresh fruit bunch yield-FFBy and average bunch weight-ABWt, the correlation was high and positive (0.78%).
Based on the similarity between groups, by the average rank and the additive values estimated by BLUP, the 10 best plants were selected both for mother plants and for a new selection cycle.
A greater efficiency was found in the evaluation of plants for the traits bunch number -BNo and fresh fruit bunch yield-FFBy and low for average bunch weight-ABWt. The evaluation period of five years proved ideal.
ACKNOWLEDGEMENTS
The oil palm breeding program of the Experimental Station Santo Domingo of INIAP. To CAPES and CNPq for granting Doctoral scholarships and research awards.
REFERENCES
References
Barros M, Pires I, Barros R, Xavier A and Cruz C. 2006. Avaliação genética de progênies de meio-irmãos de Eucalyptus grandis por meio dos procedimentos REML/BLUP e da ANOVA. Scientia Forestalis 71: 99-107.
Bhering LL, Barrera CF, Ortega D, Laviola BG, Alves AA, Rosado TB and Cruz CD. 2013. Differential response of Jatropha genotypes to different selection methods indicates that combined selection is more suited than other methods for rapid improvement of the species. Industrial Crops and Products 41: 260–265. doi: 10.1016/j.indcrop.2012.04.026
Corley RHV and Gray BS. 1976. Yield and yield components. pp. 77-86. In: Corley RHV, Hardon JJ and Wood BJ.(eds.). Oil palm research. Elsevier, Amsterdam.
Corley RH and Tinker PB. 2009. La palma de aceite. Maldonado E, Maldonado F (trad.). 4 ed. Fedepalma, Bogotá. CO. Edit. Blackwell Publishing Ltd, Oxford. 604 p.
Cros D, Denis M, Sanchez L, Cochard B, Flori A, Durand-Gasselin T, Nouy B, Omore A, Pomies V, Riou V, et al. 2015. Genomic selection prediction accuracy in a perennial crop: case study of oil palm (Elaeis Guineensis Jacq.). Theoretical and Applied Genetics 128(3): 397–410. doi: 10.1007/s00122-014-2439-z
Cruz CD and Regazzi AJ. 1994. Modelos biométricos aplicados ao melhoramento genético. Universidade Federal de Viçosa, Viçosa, MG. 390 p.
Cruz CD and Carneiro PCS. 2006. Modelos biométricos aplicados ao melhoramento genético, v.2. Segunda edición. Ed. UFV, Universidade Federal de Viçosa, Viçosa, MG. 585 p.
Fedapal. 2011. Fundación de Fomento de Exportaciones de Aceite de Palma y sus Derivados de origen Nacional. Producción, consumo y excedente de aceite de palma. Destino de exportaciones de aceite de palma ecuatoriano. Acesso em: Maio 2013.
Ferreira CBB, Lopes MTG, Lopes R, Cunha RNV, Moreira DA, Barros W S and Matiello RR. 2012. Diversidade genética molecular de progênies de dendezeiro. Pesquisa Agropecuaria Brasileira 47(3): 378-384. doi: 10.1590/s0100-204x2012000300009
Gomes M, Biondi A, Brianezi T and Glass V. 2009. O Brasil dos agrocombustíveis. Impactos das lavouras sobre a terra, o meio e a sociedade - Gordura animal, Dendê, Algodão, Pinhão-Manso, Girassol e Canola. Centro de Monitoramento dos Agrocombustíveis, Brasil. 69 p.
Henderson CR and Quaas RL. 1976. Multiple trait evaluation using relatives ́ records. Journal of Animal Science 3: 1188-1197.
Instituto Nacional de Estadística y Censo. INEC. 2010. Datos estadísticos agropecuarios. En: Sistema Estadístico Agropecuario Nacional -SEAN. Resumen ejecutivo encuesta de superficie y producción agropecuaria continua - ESPAC. Quito-Ecuador. em: http://www.ecuadorencifras.gob.ec/estadisticas-agropecuarias-2/. 14 p. consulta: noviembre 2017.
Kwong QB, Teh CK, Ong AL, Heng HY, Lee HL, Mohamed M, Low JZ, Sukganah A, Chew FT, Mayes S, et al. 2016. Development and validation of a high density SNP genotyping Array for African oil palm. Molecular Plant 9(8): 1132–1141. doi: 10.1016/j.molp.2016.04.010
Kwong QB, The CK, Ong AL, Chew FT, Mayes S, Kulaveerasingam H, Tammi M, Yeoh SH, Appleton DR and Harikrishna JA. 2017. Evaluation of methods and marker Systems in Genomic Selection of oil palm (Elaeis guineensis Jacq.). BMC Genetics 18(107): 2-9.doi: 10.1186/s12863-017-0576-5
Leung DYC and Guo Y. 2006. Transesterification of neat and used frying oil: optimization for biodiesel production. Fuel Processing Technology 87(10): 883-890. doi: 10.1016/j.fuproc.2006.06.003
Lertsathapornsuk V, Pairintra R, Aryusuk K, Krisnangkura K. 2008. Microwave assisted in continuous biodiesel production from waste frying palm oil and its performance in a 100 kW diesel generator. Fuel Processing Technology 89(12): 1330-1336. doi: 10.1016/j. fuproc.2008.05.024
Lim KC and Chan KW. 1998. Bunch component studies over the past two decades. In: Proc.1996 Int. Conf. Oil and kernel production in oil palm a global perspective. Ed. By N. Rajanaidu IE, Henson and Jalani BS. Palm oil Research Institute. Malaysia, Kuala Lumpur. pp. 133-150.
Lopes R, Cunha RNV and Resende MDV. 2012. Produção de cachos e parâmetros genéticos de híbridos de caiaué com dendezeiro. Pesquisa Agropecuária Brasileira 47(10): 1496-1503. doi: 10.1590/ s0100-204x2012001000012
Meng X, Chen G, Wang Y. 2008. Biodiesel production from waste cooking oil via alkali catalyst and its engine test. Fuel Processing Technology 89(9): 851-857. doi: 10.1016/j.fuproc.2008.02.006
Mulamba NN and Mock JJ. 1978. Improvement of yield potential of the Eto Blanco maize (Zea mays L.) population by breeding for plant traits. Egypt Journal of Genetics and Cytology 7: 40-51.
Musa BB, Saleh GB and Loong SG. 2004. Genetic variability and broad-sense heritability in two Deli-AVROS D × P breeding populations of the oil palm (Elaeis guineensis Jacq.). SABRAO. Journal of Breedingand Genetics 36: 13–22.
Noh A, Rafii MY, Mohd Din A, Kushairi A, Norziha A, Rajanaidu N, Latif MA, Malek MA. 2014. Variability and performance evaluation of introgressed Nigerian dura x deli dura oil palm progenies. Genetics and Molecular Research 13(2): 2426–2437. doi: 10.4238/2014.april.3.15
Okoye MN, Okwuagwu CO and Uguru MI. 2009. Population improvement for fresh fruit bunch yield and yield components in oil palm (Elaeis guineensis Jacq). American-Eurasian Journal of Scientific Research 4: 59-63.
Okoye MN. 2007. Population improvement and stability of bunch yield components of NIFOR second cycle oil palm hybrids. University of Nigeria, Nsukka. 33 p.
Ooi SC, Hardon JJ and Phang S. 1973. Variability in the Deli dura breeding population of the oil palm (Elaeis guineensis Jacq). I Components of bunch yield. Malaysian Agricultural Journal 49: 112-121.
Ortega DSC, Ferreira FM, Barros WS, Cruz CD, Dias LAS and Rocha RB 2008a. Selection among and within and combined selection in oil palm families derived from Dura x Dura. Ciência Rural 38(1): 65- 71. doi: 10.1590/s0103-84782008000100011
Ortega DSC, Barros WS, Ferreira FM, Dias LAS, Rocha RB and Cruz CD. 2008b. Correlation and repeatability in progenies of African oil palm. Acta Scientiarum 30(2): 197-201.x.
Ramalho MA and Lambert E. 2004. Biometria e o melhoramento de plantas na era da genômica. Revista Brasileira de Milho e Sorgo 3(2): 228-249. doi: 10.18512/1980-6477/rbms.v3n2p228-249
Rafii M, Rajanaidu N, Jalani B and Kushairi A. 2002. Performance and heritability estimations on oil palm progenies tested in different environments. Journal of Oil Palm Research 14: 15–24.
Resende MDV. 2007. Software SELEGEN-REML/BLUP, Sistema estatístico e seleção genética computarizada via modelos lineares mistos. Colombo, Embrapa Floresta. 359 p.
Resende MDV and Duarte JB. (2007) Precisão e Controle de Qualidade em Experimentos de Avaliação de Cultivares. Pesquisa Agropecuária Tropical 37: 182-194
Resende MDV. 2002. Genética Biométrica e Estatística no Melhoramento de Plantas Perenes. Brasília, Embrapa. 975 p.
Rosenquist EA. 1985. The genetic base of oil palm breeding populations. Proceedings of the International Workshop on Oil Palm Germplasm and Utilization. pp. 27-59.
Soh AC. 1994. Ranking parents by best linear unbiased prediction (BLUP) of breeding values in oil palm. Euphytica 76(1-2): 13-21. doi: 10.1007/bf00024016
Sparnaaij LD. 1969. Oil palm –Elaeis guineensis Jacquin. PP. 339387. In: Ferwerda FP and Wit F (eds.). Outlines of perennial crop breeding in the tropics . Veenman and Zonen, Wageningen.
Van der Vossen HAM. 1974. Towards more efficient selection for oil yield in the oil palm (Elaeis guineensis, Jacquin). Thesis. Centre for Agricultural Publishing and Documentation, Wageningen. 107 p.
Wong C and Bernardo R. 2008. Genomewide selection in oil palm: increasing selection gain per unit time and cost with small populations. Theoretical and Applied Genetics 116(6): 815-824. doi: 10.1007/s00122-008-0715-5
Yang Z and Xie W. 2007. Soybean oil transesterification over zinc oxide modified with alkali earth metals Fuel Processing Technology 88(6): 631-638. doi: 10.1016/j.fuproc
How to Cite
APA
ACM
ACS
ABNT
Chicago
Harvard
IEEE
MLA
Turabian
Vancouver
Download Citation
CrossRef Cited-by
1. Elivelton Alves Lustri, Walter José Siqueira, Joaquim Adelino de Azevedo Filho, Suelen Alves Vianna, Carlos Augusto Colombo. (2021). Estimates of genetic parameters for juvenile traits in macaw palm. Bragantia, 80 https://doi.org/10.1590/1678-4499.20200463.
2. Andrés Tupaz-Vera, Iván Ayala-Diaz, Carlos Felipe Barrera, Hernán Mauricio Romero. (2021). Selection of Elite dura-Type Parents to Produce Dwarf Progenies of Elaeis guineensis Using Genetic Parameters. Agronomy, 11(12), p.2581. https://doi.org/10.3390/agronomy11122581.
Dimensions
PlumX
Article abstract page views
Downloads
License
Copyright (c) 2018 Revista Facultad Nacional de Agronomía Medellín

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
The journal allows the author(s) to maintain the exploitation rights (copyright) of their articles without restrictions. The author(s) accept the distribution of their articles on the web and in paper support (25 copies per issue) under open access at local, regional, and international levels. The full paper will be included and disseminated through the Portal of Journals and Institutional Repository of the Universidad Nacional de Colombia, and in all the specialized databases that the journal considers pertinent for its indexation, to provide visibility and positioning to the article. All articles must comply with Colombian and international legislation, related to copyright.
Author Commitments
The author(s) undertake to assign the rights of printing and reprinting of the material published to the journal Revista Facultad Nacional de Agronomía Medellín. Any quotation of the articles published in the journal should be made given the respective credits to the journal and its content. In case content duplication of the journal or its partial or total publication in another language, there must be written permission of the Director.
Content Responsibility
The Faculty of Agricultural Sciences and the journal are not necessarily responsible or in solidarity with the concepts issued in the published articles, whose responsibility will be entirely the author or the authors.






















