Publicado
AISLAMIENTO E IDENTIFICACIÓN DE Lactobacillus spp. (LACTOBACILLACEAE) RESISTENTES A Cd(II) Y As(III) RECUPERADOS DE FERMENTO DE CACAO
Isolation and identification of Lactobacillus spp. (Lactobacillaceae) resistant to Cd(II) and As(III) from fermented cocoa
ISOLAMENTO E IDENTIFICAÇÃO DE Lactobacillus spp. (LACTOBACILLACEAE) RESISTENTES A Cd(II) E As(III) RECUPERADOS DE FERMENTO DE CACAU
DOI:
https://doi.org/10.15446/abc.v26n1.83677Palabras clave:
Elementos tóxicos, Fermentación, Lactobacilos, Theobroma cacao (es)Fermentation, Lactobacilli, Theobroma cacao, Toxic elements (en)
Descargas
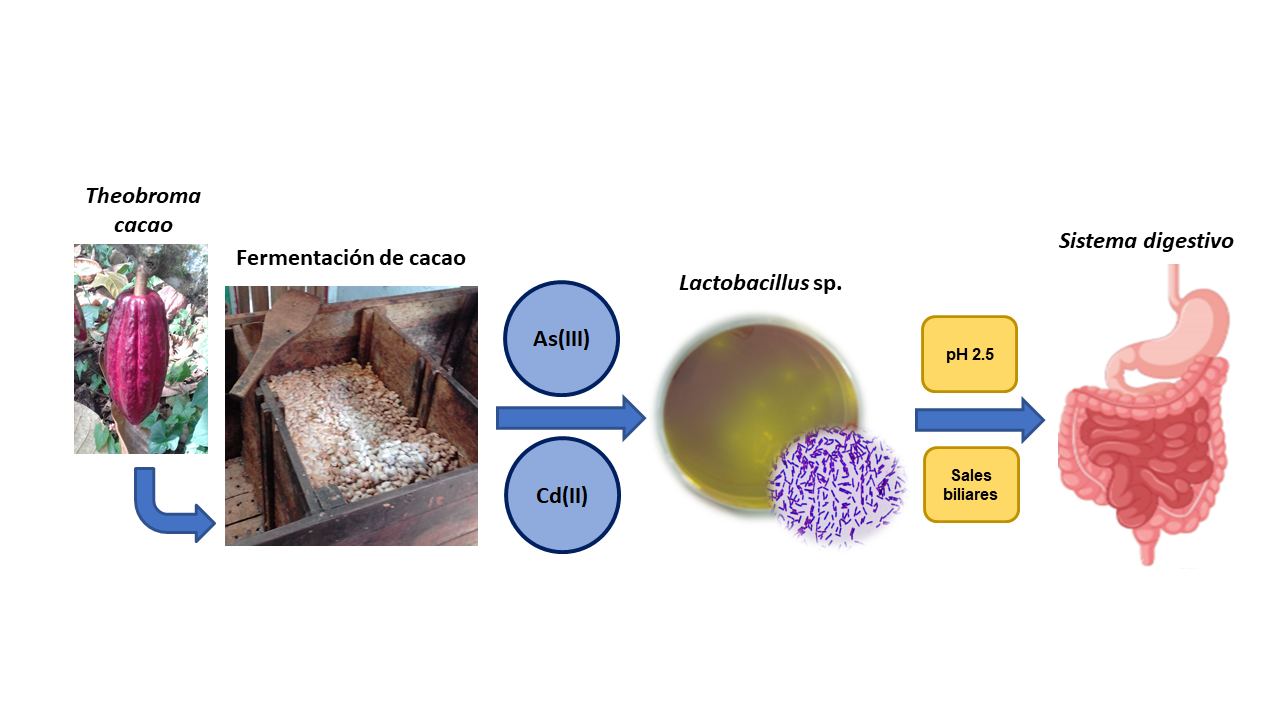
El objetivo de este estudio fue aislar e identificar a partir de cacao fermentado en Caldas Colombia, bacterias con potencial de aplicación en procesos biotecnológicos, como la detoxificación de cadmio (Cd(II)) y arsénico (As(III)) en el organismo humano. En total se recuperaron 36 aislados de los cuales se recuperaron 11 en presencia de 1,0 mg/L de Cd(II) y 25 en presencia de 0,1 mg/L de As(III). Su identificación molecular determinó que la mayoría de los aislados son del género Lactobacillus. Los ensayos de crecimiento en presencia de diferentes concentraciones de los elementos evaluados permitió determinar que gran parte de los aislamientos presentan resistencia a mayores concentraciones de As(III) (300 mg/L) que de Cd(II) (10 mg/L). En ensayos de tolerancia a la acidez (pH 2,5) se encontró que la cepa tipo Lactobacillus plantarumJCM 1055, junto con los aislamientos nativos L. plantarumA19, A26 y C16, mostraron la mayor tolerancia, por lo que se seleccionaron para evaluar su tolerancia a condiciones de salinidad. Las bacterias evaluadas mostraron crecimiento en concentraciones de hasta 4 g/L de sales biliares. Se concluye que los L. plantarumevaluados en este trabajo tienen un gran potencial para futuros ensayos en los que se busque demostrar la disminución de la bioaccesibilidad de Cd(II) y As(III) en condiciones in vitro del sistema digestivo humano debido a su resistencia a altas concentraciones de estos elementos y su tolerancia a condiciones de acidez y salinidad. Esto, junto con el reconocido potencial probiótico que tienen estos microorganismos, permitirá a futuro su uso en procesos biológicos de mitigación de Cd(II) y As(III).
The aim of this study was to isolate and identify from fermented cocoa in Caldas Colombia, bacteria with potential application in biotechnological processes such as detoxification of cadmium (Cd(II)) and arsenic (As(III)) in the human organism. In total, 36 isolates were obtained, from which 11 were recovered in the presence of 1.0 mg/L of Cd(II) and 25 in presence of 0.1 mg/L of As(III). Molecular identification showed most isolates belong to the genera Lactobacillus. Minimum inhibitory concentration assays, in presence of different concentrations of the elements, allowed to determine that the majority of isolates have resistance to higher concentration of As(III) (300 mg/L) than Cd(II) (10 mg/L). Acidity tolerance assays at pH 2.5 showed that type strain Lactobacillus plantarumJCM 1055, and native isolates L. plantarumA19, A26, and C16, presented the highest tolerance, thus they were selected to evaluate their tolerance to salinity conditions. The evaluated bacteria could grow in bile salts up to 4 g/L. It is concluded that the evaluated L. plantarumhave great potential to be used in assays in which bioaccessibility of Cd(II) and As(III) is diminished under in vitroconditions of the human digestive system, due to its resistance to high concentrations of the elements and tolerance to acidic and high bile salt conditions. These facts, together with the recognized probiotic potential of these microorganisms, may allow their future use in biological processes to mitigate Cd(II) y As(III).
Este estudo teve como objetivo isolar e identificar a partir de cacau fermentado nas Caldas, Colômbia, bactérias com potencial de aplicação em processos biotecnológicos como a desintoxicação de cadmio (Cd(II)) e arsênio (As(III)) no organismo humano. Em total foram recuperados 36 isolados, dos quais 11 foram recuperados em presença de 1 mg/L de Cd(II) e 25 de 0,1 mg/L de As(III). A sua identificação molecular indicou que a maioria dos isolados são do género Lactobacillus. Ensaios de crescimento na presencia de diferentes concentrações dos elementos avaliados permitiu determinar que a grande parte dos isolados apresentam resistência a maiores concentrações de As(III) (300 mg/L) respeito a Cd(II) (10 mg/L). Nos ensaios de tolerância a acidez (pH 2,5) foi encontrado que a estirpe tipo Lactobacillus plantarum JCM 1055, junto com os isolados nativos L. plantarum A19, A26 y C16, mostraram a maior tolerância, sendo selecionados para avaliar a sua tolerância a condiciones de salinidade. As bactérias avaliadas mostraram crescimento até 4 g/L de sais biliares. Se conclui que os L. plantarum avaliados neste trabalho mostraram um grande potencial para futuros ensaios nos que se pretende demonstrar a diminuição da bioacessibilidade de Cd(II) e As(III) em condições in vitro do sistema digestivo humano devido à sua resistência a altas concentrações destes elementos, tolerância a condiciones de acidez e salinidade. Isto, junto com o reconhecido potencial probiótico que tem estes microrganismos, poderá permitir a futuro o seu uso em processos biológicos de mitigação de cadmio e arsênio.
Referencias
Alonso DL, Latorre S, Castillo E, Brandão PFB. Environmental occurrence of arsenic in Colombia: A review. Environ Pollut. 2014;186(1):272-281. Doi: https://doi.org/10.1016/j.envpol.2013.12.009
Arias VA, Rodriguez AR, Bardos P, Naidu R. Contaminated land in Colombia: A critical review of current status and future approach for the management of contaminated sites. Sci Total Environ. 2018;618(1):199-209. Doi: https://doi.org/10.1016/j.scitotenv.2017.10.245
Bienert GP, Desguin B, Chaumont F, Hols P. Channel-mediated lactic acid transport: a novel function for aquaglyceroporins in bacteria. Biochem J. 2013;454(3):559-570. Doi: https://doi.org/10.1042/BJ20130388
Bratcher DF. Other Gram-Positive Bacilli. Long SS, Prober CG, Fischer M. Principles and Practice of Pediatric Infectious Diseases. Elsevier. 2018;5(1):786-790. Doi: https://doi.org/10.1016/B978-0-323-40181-4.00133-X.
Bhakta JN, Ohnishi K, Munekage Y, Iwasaki K. Isolation and probiotic characterization of arsenic-resistant lactic acid bacteria for uptaking arsenic. Int J Bioeng Life Sci. 2010;4(11):831-838. Doi: http://doi.org/10.5281/zenodo.1083023
Bhakta JN, Ohnishi K, Munekage Y, Iwasaki, K, Wei MQ. Characterization of lactic acid bacteria-based probiotics as potential heavy metal sorbents. J Appl Microbiol. 2012;112(6):1193-1206. Doi: https://doi.org/10.1111/j.1365-2672.2012.05284.x
Brandão PFB, Torimura M, Kurane R, Bull AT. Dereplication for biotechnology screening: PyMS analysis and PCR–RFLP–SSCP (PRS) profiling of 16S rRNA genes of marine and terrestrial actinomycetes. Appl Microbiol Biotechnol. 2002;58(1):778. Doi: https://doi.org/10.1007/s00253-001-0855-x
Chou LS, Weimer B. Isolation and characterization of acid and bile-tolerant isolates from strains of Lactobacillus acidophilus. J Dairy Sci. 1999;82(1):23-31. Doi: http://dx.doi.org/10.3168/jds.S0022-0302(99)75204-5
Das S, Dash HR. Handbook of metal-microbe interactions and bioremediation. Boca Raton: Taylor & Francis; 2017. 837 p. Doi: https://doi.org/10.1201/9781315153353
Das SC, Al-Naemi HA. Cadmium toxicity: oxidative stress, inflammation and tissue injury. ODEM. 2019;7(4):144-163. Doi: https://doi.org/10.4236/odem.2019.74012
De Angelis M, Bini L, Pallini V, Cocconcelli P, Gobbetti G. The acid-stress response in Lactobacillus sanfranciscensis CB1. Microbiol. 2001;147(1):1863-1873. Doi: https://doi.org/10.1099/00221287-147-7-1863
De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey F, et al. Bergey's Manual of Systematic Bacteriology. Vol. 3. The Firmicutes. New Delhi: Springer-Verlag; 2009. p. 465- 466. Doi: https://doi.org/10.1007/978-0-387-68489-5
De Vuyst L, Lefeber T, Papalexandratou Z, Camu N. The functional role of lactic acid bacteria in cocoa bean fermentation. In: Biotechnology of lactic acid bacteria: novel applications. United Kingdom: Wiley-Blackwell; 2010. p. 301–325. Doi: https://doi.org/10.1002/9780813820866.ch17
De Vuyst L, Weckx S. The cocoa bean fermentation process: from ecosystem analysis to starter culture development. J Appl Microbiol. 2016;121(1):5-17. Doi: https://doi.org/10.1111/jam.13045
Dohrmann AB, Tebbe CC, Kowalchuk GA, De Bruijn FJ, Head IM, Akkermans ADL, van Elsas JD. Microbial community analysis by PCR-single-strand conformation polymorphism (PCR-SSCP). In: Molecular Microbial Ecology Manual. Volumes 1 and 2. Netherlands: Springer; 2004. p 809-838. Doi: https://doi.org/10.1007/978-1-4020-2177-0_316
Evanovich E, Mattos PJSM, Guerreiro JF. Comparative genomic analysis of Lactobacillus plantarum: an overview. Int J Genomics. 2019;1(1):1-11. Doi: https://doi.org/10.1155/2019/4973214
Fahrurrozi, Rahayu EP, Nugroho IB, Lisdiyanti P. Lactic acid bacteria (LAB) isolated from fermented cocoa beans prevent the growth of model food-contaminating bacteria. AIP Conf. Proc. 2019; 2099(1):1-6. Doi: https://doi.org/10.1063/1.5098410.
Gerbino E, Carasi P, Tymczyszyn EE, Gómez-Zavaglia A. Removal of cadmium by Lactobacillus kefir as a protective tool against toxicity. J Dairy Res. 2014;81(3):280-287. Doi: https://doi.org/10.1017/S0022029914000314
Gerbino E, Mobili P, Tymczyszyn E, Fausto R, Gómez-Zavaglia A. FTIR spectroscopy structural analysis of the interaction between Lactobacillus kefir S-layers and metal ions. J Mol Struct. 2011;987(1-3):186-192. Doi: https://doi.org/10.1016/j.molstruc.2010.12.012
Grumezescu A, Holban AM. Caffeinated and cocoa based beverages. 1st Edition. In: The Science of Beverages. India: Woodhead publishing: 2019; p. 423-446.
Halttunen T, Salminen S, Tahvonen R. Rapid removal of lead and cadmium from water by specific lactic acid bacteria. Int J Food Microbiol. 2007a;14(1):30-35. Doi: https://doi.org/10.1016/j.ijfoodmicro.2006.10.040
Halttunen T, Finell M, Salminen S. Arsenic removal by native and chemically modified lactic acid bacteria. Int J Food Microbiol. 2007b;120(1-2):173–178. Doi: https://doi.org/10.1016/j.ijfoodmicro.2007.06.002
Ibrahim F, Halttunen T, Tahvonen R, Salminen S. Probiotic bacteria as potential detoxification tools: assessing their heavy metal binding isotherms. Can J Microbiol. 2006;52(9):877-885. Doi: https://doi.org/10.1139/W06-043
Kinoshita H, Sohma Y, Ohtake F, Ishida M, Kawai Y, Kitazawa H, et al. Biosorption of heavy metals by lactic acid bacteria and identification of mercury binding protein. Res Microbiol. 2013;164(7):701-709. Doi: https://doi.org/10.1016/j.resmic.2013.04.004
Koch S, Eugster-Meier E, Oberson G, Meile L, Lacroix C. Effects of strains and growth conditions on autolytic activity and survival to freezing and lyophilization of Lactobacillus delbrueckii ssp. lactis isolated from cheese Int Dairy J. 2008;18(2):187-196. Doi: https://doi.org/10.1016/j.idairyj.2007.07.009
Kumar N, Kumar V, Panwar R, Ram C. Efficacy of indigenous probiotic Lactobacillus strains to reduce cadmium bioaccessibility: An in vitro digestion model. Environ Sci Pollut Res. 2017;24(2):1241-1250. Doi: https://doi.org/10.1007/s11356-016-7779-6
Liu Y, Tang H, Lin Z, Xu P. Mechanisms of acid tolerance in bacteria and prospects in biotechnology and bioremediation. Biotechnol Adv. 2015;33(7):1484-1492. Doi: https://doi.org/10.1016/j.biotechadv.2015.06.001
Monachese MA. Sequestration of lead, cadmium and arsenic by Lactobacillus species and detoxication potential (Tesis de Maestría). Ontario: The School of Graduate and Postdoctoral Studies, 2012. p. 1-729.
Montaño-Salazar SM, Lizarazo-Marriaga J, Brandão PFB. Isolation and potential biocementation of calcite precipitation inducing bacteria from Colombian buildings. Curr Microbiol. 2018;75(3):256-265. Doi: https://doi.org/10.1007/s00284-017-1373-0
Nachlas MM, Crawford DT, Goldstein TP, Seligman AM. The histochemical demonstration of cytochrome oxidase with a new reagent for the NaDi reaction. J Histochem Cytochem. 1958;6(6):445-456. Doi: https://doi.org/10.1177/6.6.445
Papalexandratou Z, Kaasik K, Villagra Kauffmann L, Skorstengaard A, Bouillon G, Leth Espensen J, et al. Linking cocoa varieties and microbial diversity of Nicaraguan fine cocoa bean fermentations and their impact on final cocoa quality appreciation. Int J Food Microbiol. 2019;304(1):106-118. Doi: https://doi.org/10.1016/j.ijfoodmicro.2019.05.012
Reiner K. 2010. Catalase test protocol. Washington D.C.: American Society for Microbiology. p. 9.
Saito VST, Dos Santos TF, Vinderola CG, Romano C, Nicoli JR, Araújo LS, et al. Viability and resistance of lactobacilli isolated from cocoa fermentation to simulated gastrointestinal digestive steps in soy yogurt. J Food Sci. 2014;79(2):208–213. Doi: https://doi.org/10.1111/1750-3841.12326
Serra JL, Moura FG, Pereira GVM, Soccol CR, Rogez H, Darnet S. Determination of the microbial community in Amazonian cocoa bean fermentation by Illumina-based metagenomic sequencing. LWT. 2019;106(1):229-239. Doi: https://doi.org/10.1016/j.lwt.2019.02.038
Shen S, Li X, Cullen W, Weinfeld M, Le X. Arsenic Binding to Proteins. Chem Rev. 2013;113(10):7769-7792. Doi: https://doi.org/10.1021/cr300015c
Singh AL, Sarma PN. Removal of arsenic(III) from waste water using Lactobacillus acidophilus. Bioremediat J. 2010;14(2):92-97. Doi: https://doi.org/10.1080/10889861003767050
Spiro T, Stigliani W. Química Medioambiental. Segunda edición. Madrid: Pearson; 2007. 500 p.
Stephen F. Freeze-Drying and Cryopreservation of Bacteria. En: Day JG, Pennington MW, editores. Cryopreservation and Freeze-Drying Protocols. Methods in Molecular Biology. Totowa, New Jork: Humana Press; 1995. p.
Sun Z, Harris HMB, McCann A, Guo C, Argimón S, Yang X, et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun. 2015;6(1):8322. Doi: https://doi.org/10.1038/ncomms9322
van Kranenburg K, Golic N, Bongers R, Leer RJ, De Vos WM, Siezen RJ, Kleerebezem M. Functional Analysis of Three Plasmids from Lactobacillus plantarum. Appl Environ Microbiol. 2005;71(3):1223-1230. Doi: https://doi.org/10.1128/AEM.71.3.1223-1230.2005
Vanderschueren R, De Mesmaeker V, Mounicou S, Isaure MP, Doelsch E, Montalvo D, et al. The impact of fermentation on the distribution of cadmium in cacao beans. Food Res Int. 2020;127:108743. Doi: https://doi.org/10.1016/j.foodres.2019.108743
Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S Ribosomal DNA Amplification for Phylogenetic Study. J Bacteriol. 1991;173(2):697–703. Doi: http://dx.doi.org/10.1128/jb.173.2.697-703.1991
Zhai Q, Wang G, Zhao J, Liu X, Tian F, Zhang, H, Chen W. Protective effects of Lactobacillus plantarum ccfm8610 against acute cadmium toxicity in mice. Appl Environ Microbiol. 2013;79(5):1508-1515. Doi: https://doi.org/10.1128/AEM.03417-12
Zhai Q, Xiao Y, Zhao J, Tian F, Zhang H, Narbad A, Chen W. Identification of key proteins and pathways in cadmium tolerance of Lactobacillus plantarum strains by proteomic analysis. Sci Rep. 2017;7(1):1182. Doi: https://doi.org/10.1038/s41598-017-01180-x
Cómo citar
APA
ACM
ACS
ABNT
Chicago
Harvard
IEEE
MLA
Turabian
Vancouver
Descargar cita
CrossRef Cited-by
1. Camila Delgado-Vergara, Luis Marileo, Ricardo Tighe-Neira, Leovigildo Medina, Patricio Javier Barra, Paola Díaz-Navarrete, Jean Guy LeBlanc, Claudio Inostroza-Blancheteau, Sharon Viscardi. (2025). Native Lactic Acid Bacteria Enterococcus sp. and Lactobacillus sp. Improve Physiological, Biochemical and Fruit Yield of Solanum lycopersicum L. Plants Under Water Deficit. Journal of Soil Science and Plant Nutrition, 25(2), p.4897. https://doi.org/10.1007/s42729-025-02436-6.
Dimensions
PlumX
Visitas a la página del resumen del artículo
Descargas
Licencia
Derechos de autor 2020 Acta Biológica Colombiana

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-CompartirIgual 4.0.
1. La aceptación de manuscritos por parte de la revista implicará, además de su edición electrónica de acceso abierto bajo licencia Attribution-NonCommercial-ShareAlike 4.0 (CC BY NC SA), la inclusión y difusión del texto completo a través del repositorio institucional de la Universidad Nacional de Colombia y en todas aquellas bases de datos especializadas que el editor considere adecuadas para su indización con miras a incrementar la visibilidad de la revista.
2. Acta Biológica Colombiana permite a los autores archivar, descargar y compartir, la versión final publicada, así como las versiones pre-print y post-print incluyendo un encabezado con la referencia bibliográfica del articulo publicado.
3. Los autores/as podrán adoptar otros acuerdos de licencia no exclusiva de distribución de la versión de la obra publicada (p. ej.: depositarla en un archivo telemático institucional o publicarla en un volumen monográfico) siempre que se indique la publicación inicial en esta revista.
4. Se permite y recomienda a los autores/as difundir su obra a través de Internet (p. ej.: en archivos institucionales, en su página web o en redes sociales cientificas como Academia, Researchgate; Mendelay) lo cual puede producir intercambios interesantes y aumentar las citas de la obra publicada. (Véase El efecto del acceso abierto).